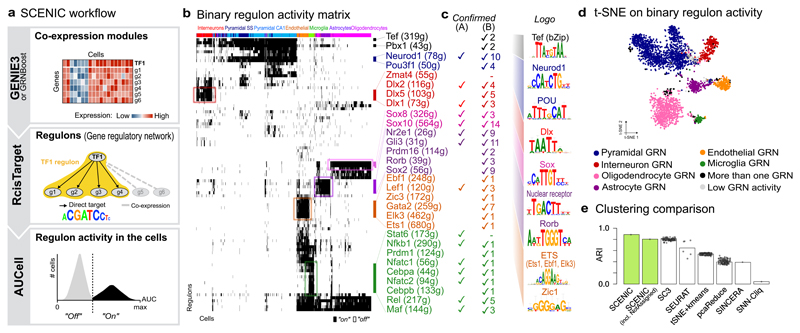
Figure 1. The SCENIC workflow and its application to the mouse brain.
(a) Co-expression modules between transcription factors and candidate target genes are inferred using GENIE3 or GRNBoost. RcisTarget identifies those modules where the regulator’s binding motif is significantly enriched across the target genes; and creates regulons with only direct targets. AUCell scores the activity of each regulon in each cell, yielding a binarized activity matrix. Cell states are based on the shared activity of regulatory subnetworks. (b) SCENIC results on the mouse brain 9; cluster labels correspond to 9; master regulators are color-matched with the cell types they control. (c) transcription factors confirmed by literature (A) or having brain phenotypes from MGI (B), and the enriched DNA motifs are shown. (d) t-SNE on the binary regulon activity matrix. Each cell is assigned the color of the most active GRN. (e) Accuracy of different clustering methods on this dataset.