Fig. 2. Efficient loxP KI at the mouse Upf1 locus using overlapping sgRNAs in zygotes.
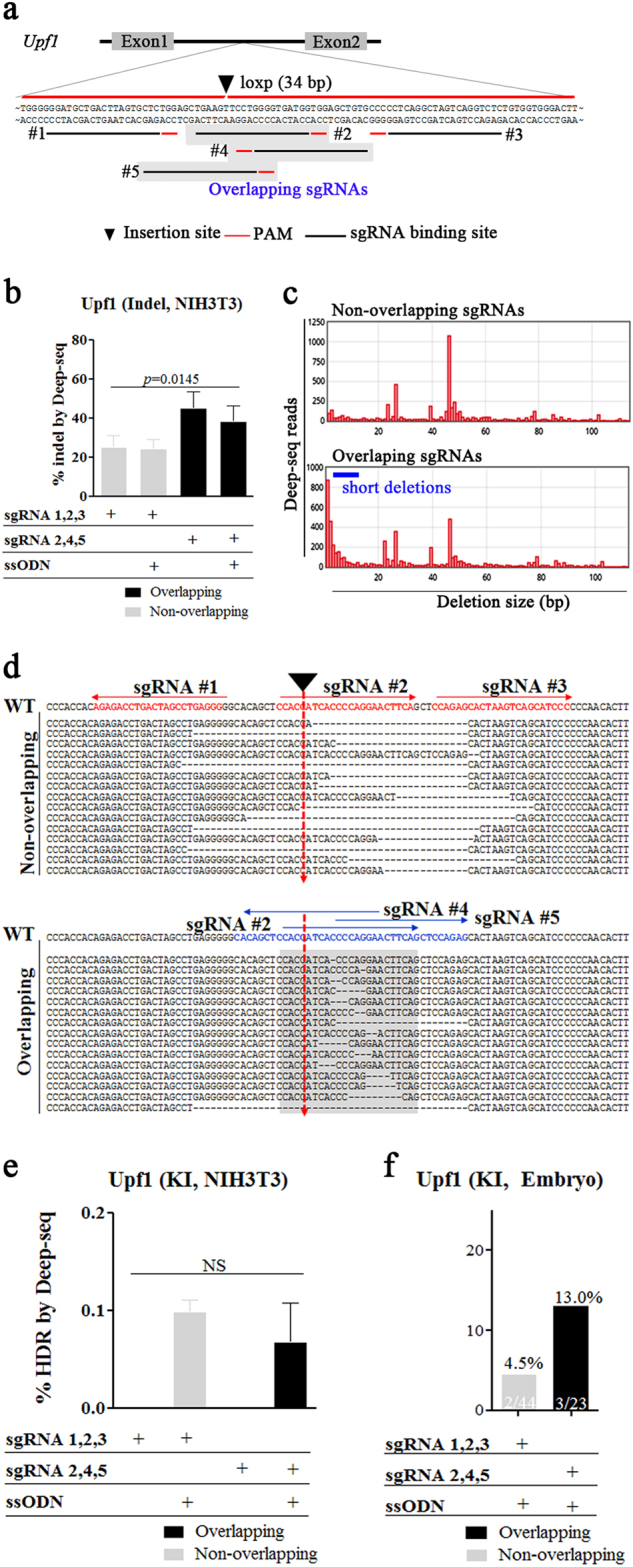
a Schematic diagram of the strategy for loxP KI using ssODNs into the Upf1 locus. For targeting of the Upf1 intron 1 locus, the two different sets of sgRNAs were used (non-overlapping; overlapping, grey box) (b–e). Targeted deep sequencing of sequencing of gDNAs from NIH-3T3 cells transfected with overlapping or non-overlapping sgRNAs with Cas9 mRNA and ssODNs. b Percentages of NHEJ-induced indels, (c) Distribution of sequence-deletion sizes associated with use of overlapping or non-overlapping sgRNAs, (d) Fifteen most frequent deletion patterns. Red and blue alphabets indicate sgRNA binding sites, red arrows indicate target sites for loxP insertion, and gray box indicate overlapping region and (e) HDR-mediated KIs. The p-values shown were calculated using Student’s t-test. f 3 days after microinjection of overlapping or non-overlapping sgRNAs with Cas9 mRNA and ssODNs, PCR-based ssODN-mediated KI efficiency was measured and is represented as bar graphs. KI (%) indicates the ratios of KI embryos derived from total blastocysts
