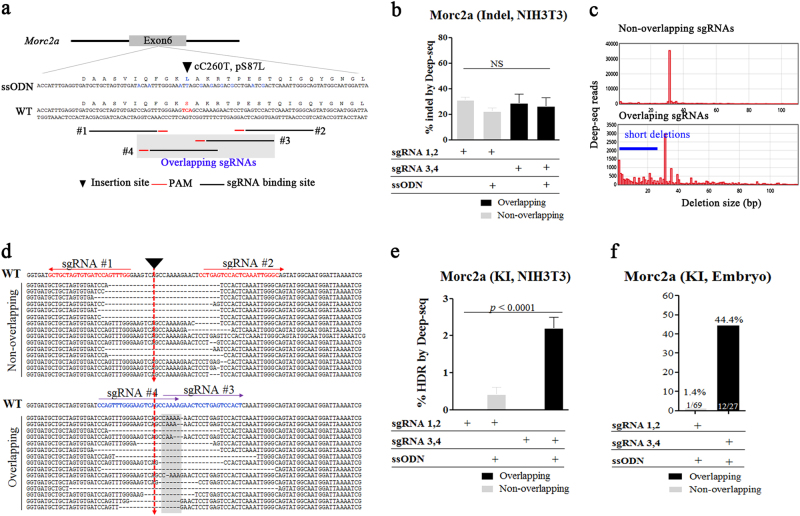
Fig. 3. Efficient incorporation of single-nucleotide substations at the mouse Morc2a locus using overlapping sgRNAs in mouse zygotes and fibroblast cell lines.
a Schematic diagram of the strategy for introducing cC260T mutations using a ssODN consisting of 60 nt of homology arms and 45 nt of the insertion sequence containing silent and targeted mutations into exon 6 of the Morc2a locus. The two different sets of sgRNAs (non-overlapping; and overlapping, grey box) are shown. Blue letters indicate nucleotide alteration for amino-acid change or silent mutation for PCR-based KI analysis (b–e). Targeted deep sequencing of gDNAs from NIH-3T3 cells transfected with overlapping or non-overlapping sgRNAs containing Cas9 mRNA and ssODN sequences. b Percentages of NHEJ-induced indels. c Distribution of sequence-deletion sizes associated with the use of overlapping or non-overlapping sgRNAs. d Fifteen most frequent deletion patterns. Red and blue alphabets indicate sgRNA binding sites, red arrows indicate target sites for loxP insertion, and gray box indicate overlapping region, and (e) HDR-mediated KIs. The p-values were calculated using Student’s t-test. f 3 days after microinjection of overlapping or non-overlapping sgRNAs with Cas9 mRNA and ssODNs, PCR-based ssODN-mediated KI efficiency was measured and is represented as bar graphs. KI (%) indicates the ratios of KI embryos derived from total blastocysts