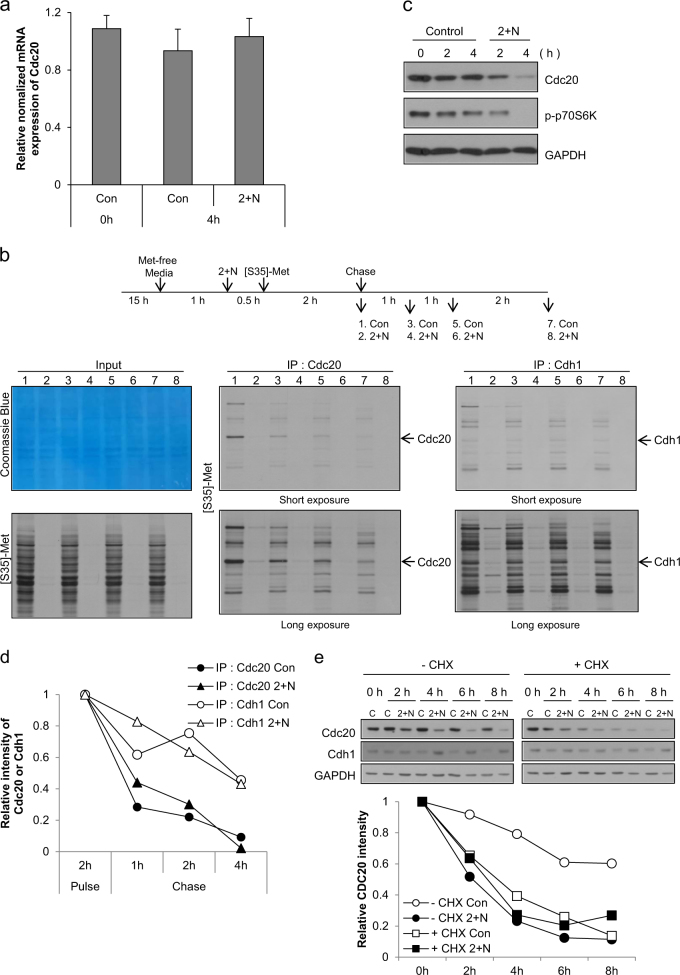
Fig. 5. Decreased protein synthesis by ATP depletion and the short half-life reduced the level of cdc20 during mitotic slippage.
a Mitotic arrested cells were treated with 2-DG and NaN3 (2 + N), and expression of Cdc20 mRNA was analyzed by real-time PCR. The results are given as the mean ± SD from three independent experiments. b Experimental design (upper panel). For protein synthesis experiments, HeLa cells grown in DMEM/high glucose were used. Cells arrested in mitosis were obtained by nocodazole treatment (100 ng/ml) for 15 h followed by shake-off, and then were incubated in methionine-free medium with nocodazole. After 1 h, cells were pre-exposed to 2-DG and NaN3 (2 + N) for 30 min. [35S]-methionine was added to the medium, and proteins were labeled for 2 h and subsequently chased by the addition of an excess of unlabeled methionine for 1 h, 2 h and 4 h. Total cell lysates and immunoprecipitated lysates using Cdc20 or Cdh1 antibody were resolved by SDS-PAGE and visualized by Coomassie Blue staining or autoradiography. c Mitotic arrested cell lysates from the indicated times after co-treatment with 2-DG and NaN3 (2 + N) were subjected to western blot analysis using antibodies for Cdc20, phospho-p70S6 kinase, and GAPDH. d Quantification of the relative intensity of Cdc20 or Cdh1 was performed by measuring the band intensity using ImageJ software. e For inhibition of protein synthesis, cells were treated with 10 μg/ml cycloheximide for 30 min prior to co-treatment with 2-DG and NaN3. Cell lysates from the indicated times after co-treatment with 2-DG and NaN3 (2 + N) were subjected to western blot analysis using antibodies for Cdc20, Cdh1, and GAPDH (upper panel). Quantification of the relative intensity of Cdc20 or Cdh1 was performed by measuring the band intensity using ImageJ software (lower panel)