Abstract
Recently, parainfluenza virus 5 (PIV5) infection has been increasingly reported in mammals. In this study, five PIV5 strains were isolated from diarrhea-affected piglets from four provinces or municipalities in China. An F-gene-based phylogenetic tree indicated that the five isolated strains were closely related to the PIV5 strain ZJQ-221 from a lesser panda in China, and the PIV5 strain 1168-1 from a dog in South Korea. The new isolates differed genetically from other pig, calf, rhesus macaque kidney cells, human, and dog PIV5 reference strains. Our study reveals the presence of PIV5 in intestinal tissue samples collected from diarrhea-affected piglets, and provides novel information regarding the epidemiology and tissue tropism of PIV5.
Keywords: diarrhea-affected piglet, isolation, parainfluenza virus 5
Parainfluenza virus 5 (PIV5) is a negative-sense, non-segmented, single-stranded RNA virus. PIV5 belongs to the genus Rubulavirus in the family Paramyxoviridae [5]. PIV5 has the smallest genome among the Paramyxoviridae family with 15,246 nucleotides, and a circular or polymorphous-shaped virion of 50–200 nm in diameter based on cryo-electron microscopy [12]. The full-length genome of PIV5 is composed of a 3′ leader region, a 5′ trailer region, and seven non-overlapping genes (NP, V/P, M, F, SH, HN and L) [3]. PIV5 was first reported in primary monkey kidney cells in 1954, and was also known as simian virus 5 (SV5) [4]. Since then, PIV5 has been frequently isolated from various susceptible hosts, including humans, pigs, dogs, cattle, cats, hamsters, guinea pigs, and lesser pandas [1, 6, 8, 14]. It has been reported that PIV5 exhibits a potential association with respiratory infection in asymptomatic hosts [8]. Until date, the data regarding PIV5 infection in digestive-system diseases has been scarce. In the current study, we provide evidence that PIV5 infections were present in the intestinal tissues of diarrhea-affected piglets from four different provinces of the People’s Republic of China.
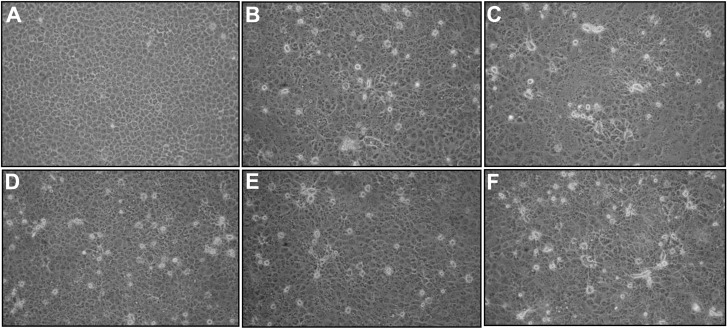
In our previous study, 137 porcine epidemic diarrhea virus (PEDV)-positive samples were collected from 18 provinces or municipalities of China in 2015 by using reverse transcription polymerase chain reaction (RT-PCR) targeting the ORF3 gene of PEDV, as described by Wang et al. [13]. The 137 samples positive for PEDV were selected for virus isolation on Vero E6 cells. The isolation was conducted according to the protocol for PEDV isolation as reported previously, with modifications [2]. Briefly, cells were cultured and maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 10 unit/ml penicillin, and 10 µg/ml streptomycin. Confluent monolayers of Vero E6 cells in 25-cm2 rectangular canted neck cell culture flasks with vent caps (Corning Inc., Corning, NY, U.S.A.) were washed twice with postinoculation medium. The postinoculation medium was DMEM, supplemented with 5 µg/ml Trypsin-EDTA (Thermo Fisher Scientific, Hudson, NH, U.S.A.). The cells were then inoculated with 0.2 ml of sample combined with 1.8 ml of postinoculation medium. After incubation at 37°C with 5% CO2 for 2 hr, the inoculum was removed and 7 ml postinoculation medium was added to each flask. Inoculated cells were incubated at 37°C with 5% CO2. When a 70% cytopathic effect (CPE) had developed, the flasks were subjected to three freeze-thaw cycles. The virus was harvested from the supernatant for further propagation or stored at −80°C. Of these PEDV-positive samples, five samples showed remarkable CPE on Vero E6 cells at the beginning of the fifth passage when compared with uninfected control cells (Fig. 1). Information regarding the five isolated strains is shown in Table 1. However, according to RT-PCR results targeting the ORF3 gene, the five isolated strains were negative for PEDV [13].
Fig. 1.
Virus-specific CPE of the five PIV5 strains on Vero E6 cells at a magnification of 200 × (fifth passage, 36 hr post infections). A, Mock-infected Vero E6 cells; B, PIV5-infected Vero E6 cells with strain HLJ2015/DP1-1/PIV5; C, PIV5-infected Vero E6 cells with strain HLJ2015/DP2-1/PIV5; D, PIV5-infected Vero E6 cells with strain SH/2015/122/PIV5; E, PIV5-infected Vero E6 cells with strain HuB/YC/2015/PIV5; F, PIV5-infected Vero E6 cells with strain JX/2015/1221/PIV5.
Table 1. Information regarding the PIV5-positive samples.
| Farm no. | Geographic location (Province) |
Strain name | Collection date | F gene (GenBank accession no.) |
|---|---|---|---|---|
| 1 | Heilongjiang | HLJ2015/DP1-1/PIV5 | Apr-2015 | KY364869 |
| 2 | Heilongjiang | HLJ2015/DP2-1/PIV5 | Apr-2015 | KY364870 |
| 3 | Shanghai | SH/2015/122/PIV5 | Dec-2015 | KY364871 |
| 4 | Hubei | HuB/YC/2015/PIV5 | Dec-2015 | KY364872 |
| 5 | Jiangxi | JX/2015/1221/PIV5 | Dec-2015 | KY364873 |
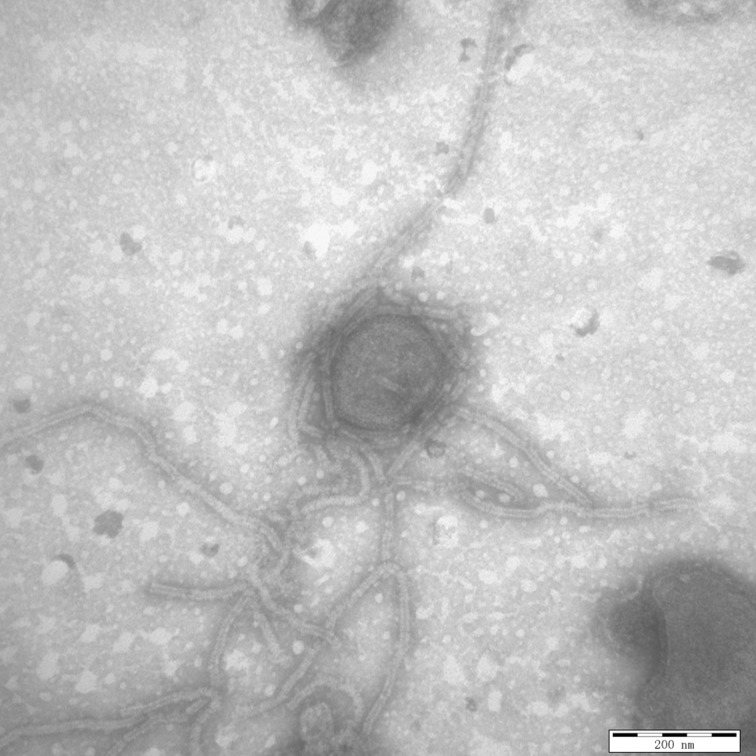
In an attempt to visually identify any viral species for the five isolated strains, transmission electron microscopy was performed using negative staining. Briefly, the viral suspension was subjected to ultracentrifugation at 30,000 × g for 30 min, and the precipitate was negatively stained with 2% phosphotungstic acid (PTA, pH 7.0). The negatively stained samples were examined using a Hitachi-7650 transmission electron microscope (Hitachi, Ltd., Tokyo, Japan). Results visually demonstrated paramyxovirus-like pleomorphic virions in the purified virus suspensions of all five isolated strains. A representative ultrastructural micrograph of the five isolated strains is shown in Fig. 2.
Fig. 2.
Transmission electron microcopy of the supernatants of the virus infected Vero E6 cells by using negative staining with phosphotungstic acid. The micrograph is representative of the five PIV5 strains identified in our study and demonstrates the PIV5 ultrastructure. (Scale bar=200 nm)
Illumina next-generation sequencing was employed to classify the five suspected strains of paramyxovirus. Briefly, complementary DNA (cDNA) synthesis of the five samples was carried out according to the protocol described by Wang and others (2016) using random hexamer primers [13]. A TruSeq RNA Sample Preparation Kit (Illumina Inc., San Diego, CA, U.S.A.) along with 150 ng of cDNA was used to construct a library according to the manufacturer’s instructions. The cDNA sample was sonicated to generate fragments less than 500 base pair (bp) by using the Covaris M220 Focused-ultrasonicator. The cDNA fragments were polished using T4 polynucleotide kinase. Adaptors were then ligated, and the samples loaded onto the Illumina HiSeq 2000 Sequencing System. Raw sequence reads were trimmed to exclude the adaptor sequence, duplicate reads, and cat genomic sequences. The trimmed reads were assembled using SOAPdenovo v2.04 software (http://soap.genomics.org.cn/), and the assembled genomes were corrected by using GapCloser v1.12 software. The prediction of viral genes was performed using the software GeneMarkS (http://topaz.gatech.edu/GeneMark/genemarks.cgi), and annotation of the predicted viruses was carried out through the nonredundant protein database (NR) using BLASTn and BLASTx. Gene annotation results were consistent with gene fragments from all five strains being identified as PIV5-associated sequences (data not shown).
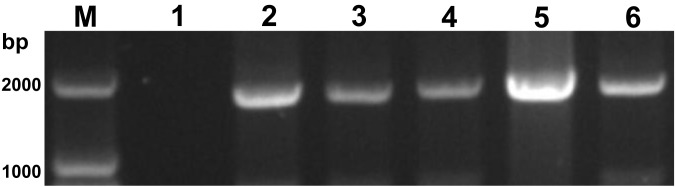
In order to further confirm the next-generation sequencing results, RT-PCR targeting the fusion (F) gene of PIV5 was carried out. The primer pairs specific for the F gene were designed according to pig PIV5 strain KNU-11 (GenBank accession no. KC852177), and included the forward primer pigF-oF (5′-AAATCATATTAAGACTAT-3′) and the reverse primer pigF-oR (5′-GTCCTATCGTTTTTTTCTTA-3′). The RNA extraction and cDNA synthesis were performed as described previously by Wang et al. [13]. The PCR amplification profile included an initial denaturation step at 95°C for 5 min; followed by 36 consecutive cycles of 94°C for 1 min, 50°C for 1 min, 72°C for 2 min; and a final extension at 72°C for 10 min. The F-gene PCR products were cloned into the pMD18-T vector, and subjected to Sanger sequencing. Sequence analysis was performed using the EditSeq program of the Lasergene DNASTAR™ v5.06 software (DNASTAR Inc., Madison, WI, U.S.A.). All nucleotide sequences generated from our study were submitted to GenBank. Results from the RT-PCR-based sequencing indicated that the five isolated strains were positive for PIV5 (Fig. 3). The five strains were named HLJ2015/DP1-1/PIV5, HLJ2015/DP2-1/PIV5, SH/2015/122/PIV5, HuB/YC/2015/PIV5 and JX/2015/1221/PIV5, respectively (Table 1). The entire F gene (1,656 bp) was successfully sequenced for each of the five PIV5 strains. A sequence comparison of the F gene among the new strains revealed 99.6–99.9% nucleotide identity and 98.9–99.6% amino-acid identity (Table 2). Comparison of the five isolates with other PIV5 reference strains from various hosts showed that they shared the highest sequence similarity with PIV5 strain ZJQ-221, which was originally isolated from a lesser panda in China. The level of sequence identity compared to ZJQ-221 ranged from 99.8–100% at the nucleotide level and 99.3–99.8% at the amino acid level (Table 2).
Fig. 3.
The electrophoretic diagram of RT-PCR amplification of the F gene from the five newly isolated PIV5 strains on 0.8% agarose gel electrophoresis with ethidium bromide staining. Lane M, DNA Marker DL2000; Lane 1, negative control; Lane 2, strain HLJ2015/DP1-1/PIV5; Lane 3, strain HLJ2015/DP2-1/PIV5; Lane 4, strain SH/2015/122/PIV5; Lane 5, strain HuB/YC/2015/PIV5; Lane 6, strain JX/2015/1221/PIV5.
Table 2. Sequence analysis of the F genes from the PIV5 strains identified in our study.
| Identity analysis | Nucleotides (%) | Amino acids (%) |
|---|---|---|
| Among the PIV5 strains identified in our study | 99.6–99.9 | 98.9–99.6 |
| Compared with other PIV5 reference strains | ||
| ZJQ-221 strain (KX100034) from lesser panda in China in 2015 | 99.8–100 | 99.3–99.8 |
| PV5-BC14 strain (KM067467) from calf in China in 2014 | 98.8–99.2 | 97.8–98.4 |
| KNU-11 strain (KC852177) from pig in South Korea in 2011 | 98.8–99.0 | 97.6–98.2 |
| SER strain (JQ743328) from pig in Germany in 1998 | 98.9–99.2 | 97.8–98.4 |
| 1186-1 strain (KC237064) from dog in South Korea in 2009 | 99.5–99.8 | 98.6–99.1 |
| CPI + strain (JQ743320) from dog in U.S.A. in 1980 | 98.0–98.2 | 96.9–97.5 |
| W3A strain (JQ743318) from rhesus macaque kidney cells in U.S.A. in 1980 | 98.4–98.7 | 97.5–98.1 |
| AGS strain (KX060176) from gastric adenocarcinoma cells in U.S.A. in 1983 | 97.4–97.6 | 96.9–97.5 |
| MIL strain (JQ743326) from human in U.K. in 1980 | 98.0–98.2 | 97.5–98.0 |
| H 221 strain (JQ743323) from dog in U.K. in 1980 | 98.1–98.2 | 97.8–98.2 |
| D277 strain (KC237065) from dog in South Korea in 2008 | 96.4–96.6 | 95.8–96.2 |
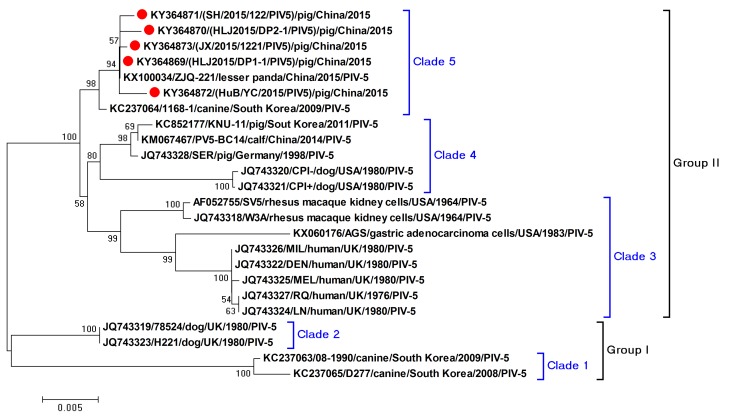
In order to trace the evolution of the five PIV5 strains, the entire F genes of the selected reference strains from the PIV5 were retrieved from the NCBI nucleotide database. Phylogenetic trees were constructed using MEGA 6.06 software with the neighbor-joining method [11]. Results indicated that the five identified PIV5 strains shared a close relationship with strain ZJQ-221, and strain 1168-1 from a dog, forming a single clade in the phylogenetic tree (Fig. 4). The five PIV5 strains, which were isolates from intestinal tissues of piglets, differed genetically from the KNU-11 strain that was isolated from a pig in South Korea. The limited data suggested that the PIV5 strains identified in China exhibited a close relationship. Additionally, the PIV5 strains exhibited genetic diversity, forming two groups and five clades, when compared with the PIV5 reference strains from various hosts (Fig. 4).
Fig. 4.
F-gene based phylogenetic tree of the five PIV5 strains using selected representative PIV5 strains as references. The red circles indicate the PIV5 strains identified in our study.
Over the past decades, PIV5 strains have been isolated from the respiratory system of different hosts [6,7,8,9, 14]. Thus far, there is little information about PIV5 infection in the intestinal tissues of swine. Currently, PEDV infection is known to cause serious losses to the pig industry in China [10]. In the current study, our initial goal was to isolate the pandemic PEDV strains from intestinal tissue samples of PEDV-affected piglets. However, five PIV5 strains were successfully isolated from the diarrhea-affected piglets in four provinces or municipalities in China. To the best of our knowledge, this is the first report of PIV5 being isolated from intestinal tissues of diarrhea-affected piglets in China. These data provide valuable information and allow for a better understanding of the epidemiology and tissue tropism of PIV5. However, further studies are necessary in order to strengthen viral surveillance in pig populations, and to clarify the potential pathogenicity of PIV5 infection.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no. 31472209), the National Key Research and Development Program of China (grant no. 2017YFD0501604-5), the Outstanding Youth Science Foundation of Heilongjiang province (grant no. JC2017007), the Heilongjiang Province Postdoctoral Science Foundation (LBH-Q16188), and the Heilongjiang Agriculture Reclamation Bureau during the 13th Five-Year Plan Period (HNK135-04-06-03).
REFERENCES
- 1.Chatziandreou N., Stock N., Young D., Andrejeva J., Hagmaier K., McGeoch D. J., Randall R. E.2004. Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5). J. Gen. Virol. 85: 3007–3016. doi: 10.1099/vir.0.80200-0 [DOI] [PubMed] [Google Scholar]
- 2.Chen Q., Li G., Stasko J., Thomas J. T., Stensland W. R., Pillatzki A. E., Gauger P. C., Schwartz K. J., Madson D., Yoon K. J., Stevenson G. W., Burrough E. R., Harmon K. M., Main R. G., Zhang J.2014. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 52: 234–243. doi: 10.1128/JCM.02820-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henrickson K. J.2003. Parainfluenza viruses. Clin. Microbiol. Rev. 16: 242–264. doi: 10.1128/CMR.16.2.242-264.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsiung G. D.1972. Parainfluenza-5 virus. Infection of man and animal. Prog. Med. Virol. 14: 241–274. [PubMed] [Google Scholar]
- 5.Lamb R. A., Parks G. D.2007. Fields virology. pp. 1449–1496. In: Paramyxoviridae: the Viruses and Their Replication, 5th ed. (Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B. and Straus, S. E. eds.), Lippincott Williams and Wilkins, Philadelphia. [Google Scholar]
- 6.Lee Y. N., Lee C.2013. Complete genome sequence of a novel porcine parainfluenza virus 5 isolate in Korea. Arch. Virol. 158: 1765–1772. doi: 10.1007/s00705-013-1770-z [DOI] [PubMed] [Google Scholar]
- 7.Liu C., Li X., Zhang J., Yang L., Li F., Deng J., Tan F., Sun M., Liu Y., Tian K.2017. Isolation and genomic characterization of a canine parainfluenza virus type 5 strain in China. Arch. Virol. 162: 2337–2344. doi: 10.1007/s00705-017-3387-0 [DOI] [PubMed] [Google Scholar]
- 8.Liu Y., Li N., Zhang S., Zhang F., Lian H., Hu R.2015. Parainfluenza virus 5 as possible cause of severe respiratory disease in calves, China. Emerg. Infect. Dis. 21: 2242–2244. doi: 10.3201/eid2112.141111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oem J. K., Kim S. H., Kim Y. H., Lee M. H., Lee K. K.2015. Molecular characteristics of canine parainfluenza viruses type 5 (CPIV-5) isolated in Korea. Can. J. Vet. Res. 79: 64–67. [PMC free article] [PubMed] [Google Scholar]
- 10.Sun D., Wang X., Wei S., Chen J., Feng L.2016. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: a mini-review. J. Vet. Med. Sci. 78: 355–363. doi: 10.1292/jvms.15-0446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terrier O., Rolland J. P., Rosa-Calatrava M., Lina B., Thomas D., Moules V.2009. Parainfluenza virus type 5 (PIV-5) morphology revealed by cryo-electron microscopy. Virus Res. 142: 200–203. doi: 10.1016/j.virusres.2008.12.017 [DOI] [PubMed] [Google Scholar]
- 13.Wang E., Guo D., Li C., Wei S., Wang Z., Liu Q., Zhang B., Kong F., Feng L., Sun D.2016. Molecular characterization of the orf3 and s1 genes of porcine epidemic diarrhea virus non s-indel strains in seven regions of china, 2015. PLoS One 11: e0160561. doi: 10.1371/journal.pone.0160561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai J. Q., Zhai S. L., Lin T., Liu J. K., Wang H. X., Li B., Zhang H., Zou S. Z., Zhou X., Wu M. F., Chen W., Luo M. L.2017. First complete genome sequence of parainfluenza virus 5 isolated from lesser panda. Arch. Virol. 162: 1413–1418. doi: 10.1007/s00705-017-3245-0 [DOI] [PMC free article] [PubMed] [Google Scholar]