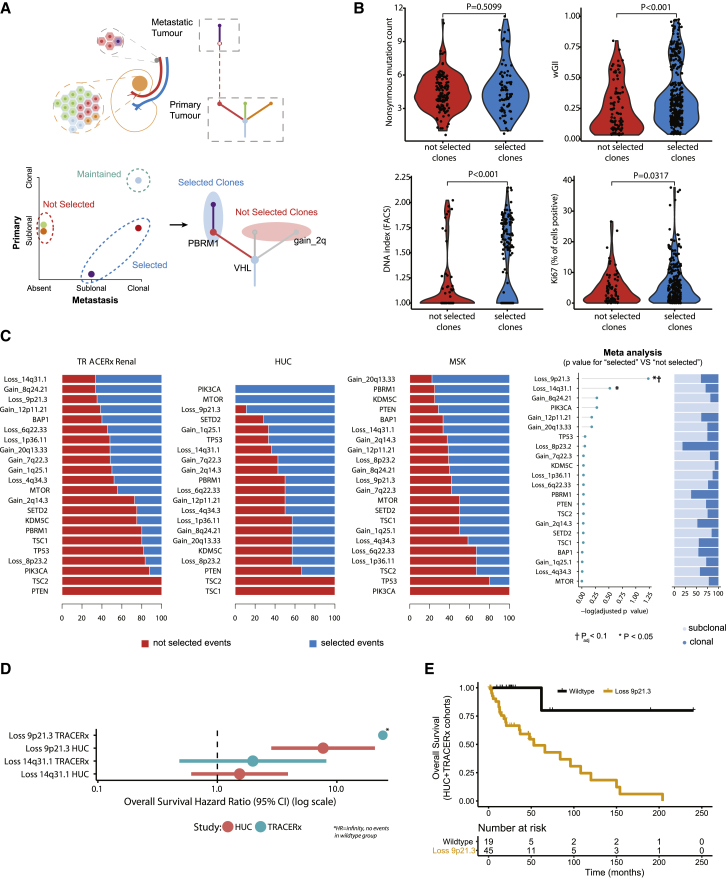
Figure 2.
Characterization of a Metastasizing Clone(s)
(A) Illustration of the method used to categorize tumor clones.
(B) Four violin plots summarizing (starting at the top left and working clockwise): (1) non-synonymous mutation count, (2) wGII, (3) ploidy, and (4) Ki67. Values are compared between tumor clones “not selected” and “selected” in metastasis, with all region/clone values plotted per tumor (excluding MRCA “maintained” clones; see the STAR Methods). A linear mixed effects (LMEs) model was used to determine significance, to account for the non-independence of multiple observations from individual tumors.
(C) For each driver event the proportion of times it was observed in “not selected” and “selected” clones for TRACERx, HUC, and MSK cohorts. The far-right panel shows the log10 p value for each event for enrichment in “selected” versus “not selected” clones. Testing was performed using a binomial test with meta-analysis conducted using Fisher’s method of combining p values from independent tests. p values are corrected for multiple testing using Benjamini-Hochberg procedure.
(D) Overall survival hazard ratios for events with p < 0.05 in the analysis in (C). Data are shown for TRACERx and HUC cohorts separately, with the circle representing the hazard ratio value and the lines corresponding to the 95% confidence interval estimate.
(E) Overall survival results for TRACERx and HUC cohorts (combined), split into two groups based on SCNA status at chr 9p21.3 (either copy-number loss at chr 9p21.3 or normal wild-type copy number) in the metastasizing clone.