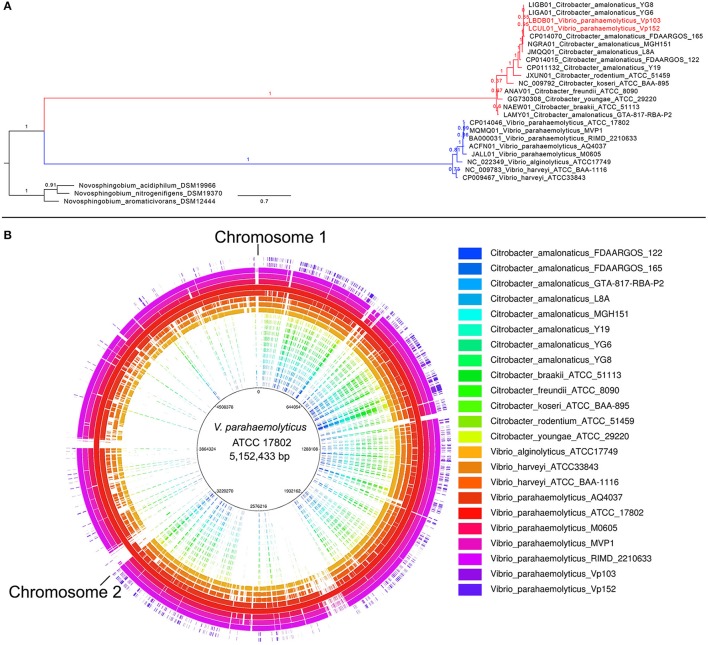
Figure 1.
Potential misidentification of strain VP152 to the genus Vibrio as revealed by phylogenomic analysis and whole genome nucleotide comparison with V. parahaemolyticus ATCC 17802T. (A) Maximum likelihood tree constructed from the concatenated alignment of 400 universal proteins. The tree was rooted with members of the genus Novosphingobium as the outgroup. Node values indicate SH-like local branch support as implemented in FastTree2 and scale bar indicates the number of amino acid changes per site. (B) Whole genome comparisons of various Vibrio and Citrobacter strains against the complete genome of V. parahaemolyticus ATCC 17802T. Regions exhibiting significant homology (BLASTN with an E-value cut-off of 1E−7) were colored based on strain label.