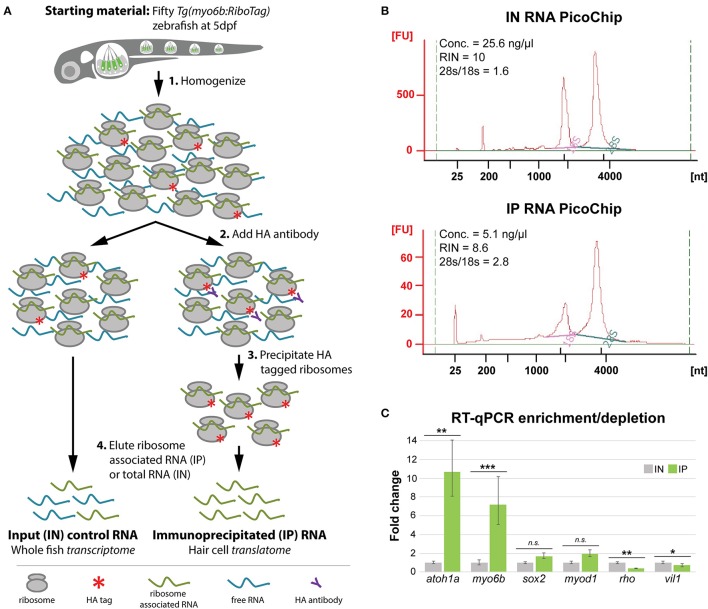
Figure 2.
HC ribosome immunoprecipitation pulls down HC-specific transcripts. (A) Explanation of the HA-tagged ribosome immunoprecipitation protocol (see Methods for more information). (B) Agilent Bioanalyzer PicoChip outputs showing that input (IN) and immunoprecipitated (IP) RNA quality is comparable between sample types despite reduced levels of 18S rRNA in the IP samples. This observation is consistent with previous zebrafish ribosome immunoprecipitation protocols (Tryon et al., 2013). (C) RT-qPCR results showing that transcripts for known HC-expressed genes such as atoh1a and myo6b are significantly enriched by HA-tagged ribosome immunoprecipitation, whereas transcripts for genes not specifically expressed in HCs are either not significantly enrichened (sox2, myod1) or depleted (rho, vil1). Error bars represent fold change ± standard deviation, and statistical significance was assessed by two-tailed Welch's t-test (n = 10). *p-value < 0.05, **p-value < 0.01, ***p-value < 0.001.