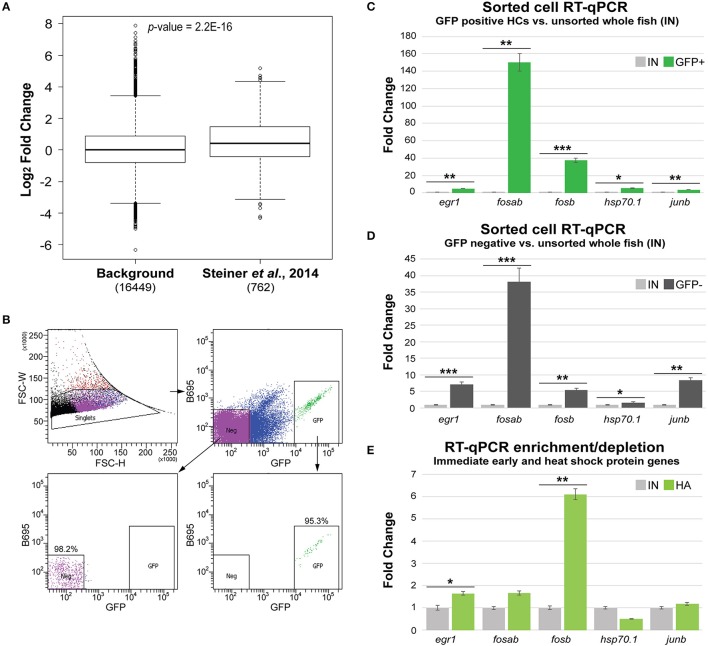
Figure 4.
Tg(myo6b:RiboTag) immunoprecipitation avoids gene expression changes from cell sorting. (A) Sets of genes found to be enriched in sorted HCs from Steiner et al. (2014) have significantly higher fold change enrichment in the IP samples compared to remaining expressed genes (background). Significance was determined by Wilcoxon's test. (B) Cell sorting experiment utilizing the Tg(myo6b:RiboTag) model. Populations of both GFP positive (GFP+) and GFP negative (Neg, GFP–) were collected via FACS based on GFP expression. Post sort analysis shows high purity of negative and positive populations (98.2 and 95.3%, respectively). (C–E) RT-qPCR analysis of zebrafish homologs of immediate early and heat shock protein encoding genes in sorted HCs (GFP+, C), sorted non-HCs (GFP–, D), and immunoprecipitated RNA (IP, C) vs. RNA extracted from whole, non-dissociated larvae (IN). Error bars represent fold change ± standard deviation. Statistical significance was assessed by two-tailed Welch's t-test (n = 3). *p-value < 0.05, **p-value < 0.01, ***p-value < 0.001.