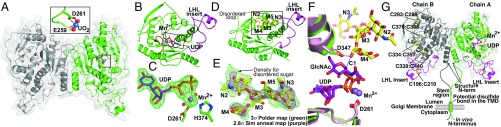
Fig. 2.
The MGAT2 structure, substrate binding, proposed mechanism, and proposed transmembrane structure. (A) The MGAT2:UO2 asymmetric unit (monomers green and gray). (Inset) The interactions between DxD motif residues (green sticks) and the bound UO2 (U and O atoms are blue and red spheres, respectively). (B) MGAT2:UDP structure (Mn2+, purple sphere; UDP, purple sticks) with LHL insert (magenta) and residues ordered following Mn2+ binding (Loop 375–387, orange). Box indicates the region for the difference map in C. (C) A 1.6-Å difference density map (Fo − Fc) of the donor analog contoured at 3σ before modeling UDP and Mn2+. (D) The MGAT2:Acc complex with acceptor bound (yellow sticks) and the LHL181–224 loop (magenta) indicated. Box indicates the region for the difference map shown in E. (E) A 2.8-Å difference density map (Fo − Fc) contoured at 3σ (purple) was calculated after omitting acceptor (yellow sticks) and subjecting the model to simulated annealing. An unbiased Polder map (24) contoured at 3σ (green) illustrates weak density that would otherwise be obscured by bulk solvent (SI Appendix, SI Materials and Methods). (F) UDP-GlcNAc modeled in MGAT2:UDP (green) using the MGAT1:UDP-GlcNAc complex (pink, Protein Data Bank ID code 1FOA) (12). Ligands are colored as in B–E. Proposed mechanism involves the catalytic base D347 deprotonating the O2 hydroxyl of M4, which attacks the C1 of the UDP-GlcNAc donor (blue dashed lines). (G) The proposed membrane-bound form of the MGAT2:UDP dimer is shown. UDP (purple sticks), Mn2+ (slate spheres), disulfide bonds (yellow spheres), LHL181–224 loop (magenta), and diagrammatic representation of the “stem region” (green and gray lines) and NH2-terminal transmembrane anchor for the full-length enzyme found in vivo.