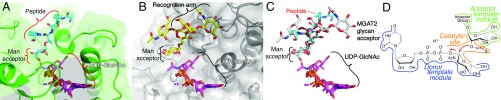
Fig. 4.
Comparison of the MGAT2 and POMGNT1 acceptor complex structures. (A) The POMGNTI:UDP:glycopeptide acceptor complex (Protein Data Bank ID code 5GGI) (25), depicted as a green surface with the glycopeptide as cyan sticks and the (B) MGAT2:acceptor and MGAT2:UDP complexes (gray surface, acceptor as yellow sticks) were aligned. Peptide and Man acceptor residue (POMGNT1), recognition arm and Man acceptor residue (MGAT2), bound UDP and modeled UDP-GlcNAc (purple sticks, Mn2+ as small magenta spheres) are indicated by brackets. (C) Overlay of the acceptor and donor structures from A and B. (D) Generalized model for GT active-site modules, where the “Donor template module” facilitates selective sugar-nucleotide donor binding, the “Acceptor template module” selectively binds the acceptor to appropriately position the hydroxyl nucleophile, and the “Catalytic site” facilitates base-catalyzed nucleophilic attack by the acceptor to the C1 of the sugar donor, in this case using an inverting catalytic mechanism.