Significance
Diatoms contribute considerably to the primary productivity of aquatic ecosystems. In diatoms, the principal storage polysaccharide is chrysolaminarin, consisting of branched β-1,3-glucans. The β-1,3-glucans are distributed widely in organisms and, besides starch and glycogen, represent major storage polysaccharides in nature. However, the synthesis pathway of storage β-1,3-glucans is still unclear. We report the localization of the glucan synthase, and thus the elongation of the glucan polymer, in the tonoplast membrane of the diatom Phaeodactylum tricornutum. Reduced expression of this glucan synthase results in various cellular effects. Our data indicate that the capacity of vacuolar polysaccharide storage substantially influences the photosynthetic performance of the plastid.
Keywords: chrysolaminarin; β-1,3-glucan synthase; photosynthesis; thylakoids; vacuole
Abstract
The β-1,3-glucan chrysolaminarin is the main storage polysaccharide of diatoms. In contrast to plants and green algae, diatoms and most other algal groups do not accumulate storage polysaccharides in their plastids. The diatom Phaeodactylum tricornutum possesses only a single gene encoding a putative β-1,3-glucan synthase (PtBGS). Here, we characterize this enzyme by expressing GFP fusion proteins in P. tricornutum and by creating and investigating corresponding gene silencing mutants. We demonstrate that PtBGS is a vacuolar protein located in the tonoplast. Metabolite analyses of two mutant strains with reduced amounts of PtBGS reveal a reduction in their chrysolaminarin content and an increase of soluble sugars and lipids. This indicates that carbohydrates are shunted into alternative pathways when chrysolaminarin production is impaired. The mutant strains show reduced growth and lower photosynthetic capacities, while possessing higher photoprotective abilities than WT cells. Interestingly, a strong reduction in PtBGS expression also results in aberrations of the usually very regular thylakoid membrane patterns, including increased thylakoid thickness, reduced numbers of thylakoids per plastid, and increased numbers of lamellae per thylakoid stack. Our data demonstrate the complex intertwinement of carbohydrate storage in the vacuoles with carbohydrate metabolism, photosynthetic homeostasis, and plastid morphology.
Diatoms are unicellular photoautotrophic eukaryotes contributing significantly to the primary production in global or oceanic habitats (1). The principal storage polysaccharide of diatoms is chrysolaminarin (2, 3), which can constitute up to 80% of organic dry weight in certain growth phases (4). This linear, noncrystalline β-1,3-glucan with branching β-1,3-glucan units added at the C-6 position is stored in vacuoles (5). The β-1,3-glucans not only represent typical metabolites of stramenopiles (to which diatoms belong as well as, e.g., brown algae or eustigmatophytes) but are also generally widely distributed among living organisms (6), in which, besides serving as storage compounds, they are involved in various processes, such as cell division, cell wall formation, or colonization (7, 8). The β-1,3-glucan is synthesized from UDP-glucose (UDPG) (9). So far, the enzyme responsible for chrysolaminarin synthesis has not been identified with experimental confidence. A gene putatively involved in chrysolaminarin synthesis has been annotated as a β-1,3-glucan synthase (BGS) in Phaeodactylum tricornutum [Phatr2 database, protein ID 55327; Joint Genome Institute (JGI)] (10). This gene shows a diel expression pattern (11), while its homolog in the diatom Thalassiosira pseudonana (Tpbgs) is up-regulated after silicon starvation (12). In that study, the authors attempted to silence this gene via an antisense approach (12). They could show a reduction of the chrysolaminarin content, but were not able to demonstrate that, and if so, to what extent the amount of TpBGS protein is reduced in these transformants (12).
In land plants and in green algae, the storage carbohydrates (starch, an α-1,4 glucan) are located in the plastids only, while in other photosynthetic organisms (e.g., glaucophytes, red algae, euglenida, chlorarachniophytes, haptophytes, stramenopiles, apicomplexans, dinoflagellates), they are found either in vacuoles or the cytosol (13). Thus, plastids serving as the storage compartment for carbohydrates seem to be more the exception than the rule. Recently, it has been reported that a BGS in Euglena is responsible for the biosynthesis of paramylon, which is a conglomerate of linear β-1,3-glucan chains (14). In plants/green algae, disrupting the starch synthesis affects the cellular energy balance and results in photochemical damage (15, 16). However, in diatoms, the transport of carbohydrates from the plastid (where photosynthesis takes place) to the vacuole (where chrysolaminarin is stored) and the coordination between the photosynthetic system (the source of carbon flux) and the polysaccharide biosynthetic and mobilization pathways (the sink of carbon flux) still are not understood.
Here, we experimentally validate the predicted BGS from P. tricornutum (PtBGS) to be the chrysolaminarin synthase by producing and characterizing RNAi-PtBGS mutants. We demonstrate that PtBGS is a tonoplast-located protein. We furthermore show that extraplastidic chrysolaminarin production at the tonoplast is not only strongly influential on the carbohydrate metabolism but also apparently connected in a complex intertwinement to photosynthetic homeostasis and plastid morphology.
Results
Stramenopile Glucan Synthases Are Related to the Respective Enzymes of Rhizaria and Haptophytes.
Basically, two overlapping gene models are provided by the genome database of P. tricornutum (Phatr2 database; JGI). We have decided to work with JGI protein ID 56808 instead of JGI protein ID 55327, which is obviously supported by the EST tracks that indicate the absence of a predicted intron in JGI protein ID 56808 (Fig. S1). The deduced polypeptide of PtBGS contains 23 putative transmembrane helices, which form hydrophobic clusters upstream and downstream of the FKS1_domain1 and at the C terminus of the BGS catalytic domain. The predicted structural organization of PtBGS basically resembles that described for yeast or plant BGSs (17, 18) (Fig. S2), suggesting that PtBGS also represents a membrane-localized protein acting as a glucan synthase by adding glucose monomers to a glucan chain on one side of the membrane while transporting the chain across the membrane. Interestingly, the diatom BGS protein contains an N-terminal cluster of transmembrane helices, which is lacking in the orthologs from yeast or plants (Fig. S2).
BGSs are only found in fungi (here involved in cell wall synthesis); viridiplantae (as callose synthases); and organisms that evolved by incorporating a eukaryotic alga (via secondary endosymbiosis), like the stramenopiles, for both synthesis of the cell wall and of storage compounds (19). Our phylogenetic analysis clearly shows that all BGSs are related to each other. These analyses also demonstrate that eukaryotic glucan synthases (BGSs) can be divided into two distinct groups (6) (Fig. S3). While the first group consists of glucan synthases from fungi only, the second group comprises individual subgroups, including synthases from euglenida, stramenopiles, alveolates, rhizaria, haptophytes, and viridiplantae. This second group is clearly separated from the fungi clade as reported earlier (6), indicating an early evolutionary separation (Fig. S3). In addition, both “glucan synthase” and “FKS1_dom1” domains are present in the two groups, which suggests that this domain organization might have already evolved in their common ancestor. Intriguingly, some members of group II enzymes do possess additional domains. For instance, streptophyte BGSs contain an additional domain (Vta1). Diatom BGSs, including PtBGS, constitute a well-supported subcluster within the stramenopile BGS subgroup. Interestingly, this stramenopile BGS subgroup and BGSs from rhizaria form sister groups that cluster together with haptophyte BGS isoforms, suggesting a close relation of the corresponding enzymes (Fig. S3). In contrast to photosynthetic stramenopiles, most of the oomycetes possess two different groups of BGSs, which cluster with the synthases of plants or with proteins from stramenopiles (Fig. S3). Stramenopile BGSs are related to enzymes from streptophyta and clearly separated from those of chlorophyta (bootstrap value = 100) (Fig. S3), indicating the same origin for stramenopile and streptophyte sequences.
PtBGS Is Located in Tonoplast.
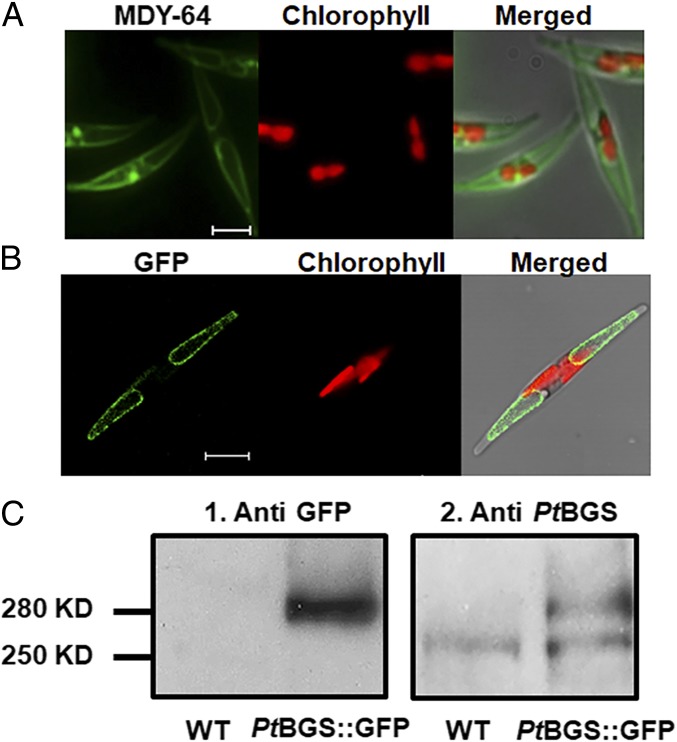
To examine the subcellular location of the enzyme, we performed a GFP-based targeting analysis with the egfp gene being C-terminally fused to the Ptbgs gene. P. tricornutum transformants expressing PtBGS::GFP show a fluorescence pattern (Fig. 1B and Fig. S4) consistent with that of vacuoles when labeled with the tonoplast maker MDY-64 (Fig. 1A), indicating that PtBGS resides in the tonoplast.
Fig. 1.
Localization of PtBGS in P. tricornutum. (A) MDY-64 stain of vacuoles in P. tricornutum. MDY-64 fluorescence (Left), chlorophyll fluorescence (Center), and a merged image (Right) are shown. (Scale bar: 5 μm.) (B) PtBGS::GFP fusion protein expressed in P. tricornutum. GFP fluorescence (Left), chlorophyll fluorescence (Center), and a merged image (Right) are shown. (Scale bar: 5 μm.) (C) Immunodetection of PtBGS::GFP in P. tricornutum. Western blots of proteins from WT and PtBGS::GFP lines (PtBGS::GFP) decorated with the antibodies against GFP (1) or PtBGS (2) are shown.
The deduced PtBGS has a calculated molecular mass of 242 kDa. Because of the large size of the Ptbgs::gfp gene construct, we could not exclude the possibility that the DNA fragment might break during the biolistic transformation process, potentially translating into shorter, truncated gene products. To exclude this possibility for the studied strains, we performed immunoblots. The utilized GFP antisera labeled a band of 280 kDa in the PtBGS::GFP expression cell line that corresponds to the calculated size of the PtBGS::GFP fusion protein. There is no such band visible in WT cellular extracts (Fig. 1 C1). Antisera that specifically label diatom PtBGS instead detect both the endogenous PtBGS (250 kDa) and the PtBGS::GFP fusion protein (280 kDa) in the expression line, while only the PtBGS band is detected in the WT cells (Fig. 1 C2). These results confirm that the transformants synthesize the fusion construct in full length.
Reduced PtBGS Expression Affects Carbohydrate Metabolism and Growth of P. tricornutum.
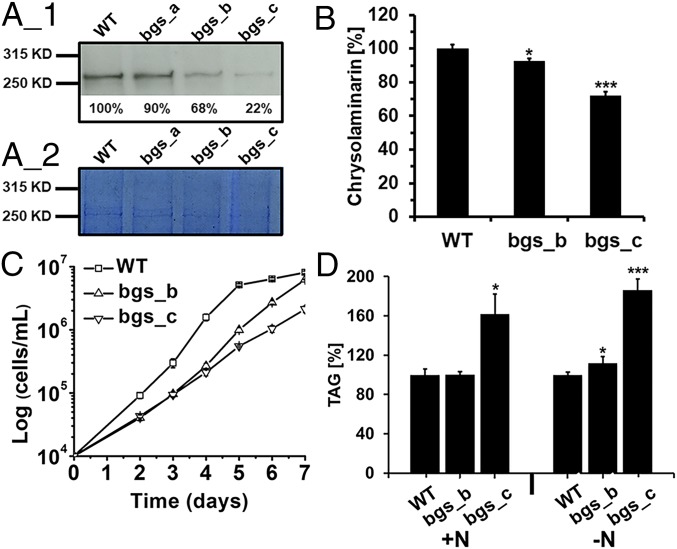
For RNAi-based posttranscriptional gene silencing of PtBGS, we transformed P. tricornutum with plasmids containing inverted repeats of the PtBGS sequence and obtained three silencing lines from three independent transformations. Western blots with BGS-specific antisera indicate that these mutant lines, named bgs_a, bgs_b, and bgs_c, exhibit 90%, 68%, and 22%, respectively, of the amount of PtBGS that is found in the WT cells (considered as 100%; Fig. 2A). Because of the low degree of PtBGS reduction, bgs_a was omitted from further analyses. Quantification of the extracted β-1,3-glucan pool indicates that chrysolaminarin production in bgs_b is slightly impaired and that chrysolaminarin production in bgs_c is considerably impaired compared with WT cells (Fig. 2B). Limited chrysolaminarin synthesis does not cause significant accumulation of the direct nucleotide sugar precursor, as reflected by identical UDPG levels in WT and bgs_c cells (Fig. S5A). However, bgs_c shows a slight but significant increase (∼1.6-fold) in soluble sugars (Fig. S5 B and C). Generally, bgs_c also exhibits higher triacylglycerol (TAG) levels than WT cells and bgs_b (Fig. 2D). Moreover, compared with WT cells, TAG content in bgs_b is identical under standard conditions but higher under nitrogen deprivation. This is indicative of a stimulated lipid biosynthesis in the mutants probably due to a substrate surplus, suggesting that accumulating precursors of chrysolaminarin synthesis (phosphorylated sugars) are shuttled into alternative pathways.
Fig. 2.
Effects of reduced PtBGS expression on chrysolaminarin growth and lipid synthesis. (A) Analysis of PtBGS protein levels in WT and Ptbgs mutants of P. tricornutum: Western blot using antisera directed against PtBGS. (A_1) Calculated expression levels are indicated, and WT is set to 100%. (A_2) Identical amounts of total protein were used for SDS/PAGE. (B) Contents of chrysolaminarin in WT and bgs mutants. Growth (C) and TAG content (D) are shown. Cells were harvested at exponential phase (around 3–4 × 106 cells per milliliter) for performing Western blot analysis and measuring chrysolaminarin. Concentrations of the WT were set to 100%, and corresponding values of bgs_b and bgs_c were calculated accordingly. SEs are indicated. Significance of the differences was evaluated by two-sided Student’s t tests. P values are indicated (***P < 0. 001; *P < 0.05).
When cultured under constant light, growth of bgs_b, and especially of bgs_c, is markedly impaired (Fig. 2C and Fig. S6). However, when cultured under either 8 h/16 h of light/dark, or 4 h/20 h of light/dark, the growth inhibition caused by silencing Ptbgs is no longer apparent (Fig. S6). It has been reported that RNAi knockdown may elicit an overall reduced growth phenotype that might not be directly linked to the silenced gene (20). However, the similar growth rates at shorter illumination periods imply that the retarded growth of the Ptbgs mutants under constant light is indeed caused by the reduction of PtBGS and, accordingly, a lower amount of chrysolaminarin.
Reduced PtBGS Expression Affects Photosynthesis of P. tricornutum.
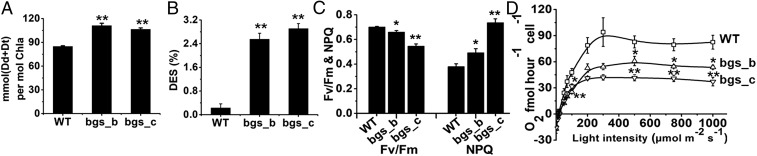
Alterations in cellular metabolism of diatoms can influence their photosynthetic performance (21, 22). Diadinoxanthin (Dd) and diatoxanthin (Dt) represent pigments of the xanthophyll cycle in diatoms. Elevated thylakoidal proton gradients (usually generated under high light conditions) activate Dd de-epoxidation into Dt, whereas Dt epoxidation into Dd occurs in the absence of a proton gradient (usually under low light or darkness) (23). Dt contributes to photoprotection as an antioxidant and via non-photochemical quenching (NPQ) (reviewed in ref. 24). Ptbgs mutants exhibit both significantly higher amounts (Fig. 3A and Table S1) and an increased de-epoxidation state (Fig. 3B) of the xanthophyll cycle pigments compared with WT cells. This indicates that the Ptbgs knockdown mutants apparently suffer from light stress even under the low light cultivation conditions. Moreover, the mutant lines show enhanced NPQ capacities and a lower maximum photosystem II (PSII) quantum yield (Fv/Fm) compared with WT cells (Fig. 3C). Our data suggest a correlation between the level of PtBGS expression and photosynthetic capacities: With decreasing PtBGS amounts (WT > bgs_b > bgs_c), Fv/Fm also decreases, whereas NPQ rises (Fig. 3C).
Fig. 3.
Effects of reduced PtBGS expression on photosynthetic properties. (A) Xanthophyll cycle pigments (Dd and Dt), normalized to chlorophyll a (Chla). (B) De-epoxidation state (DES) of the xanthophyll cycle pigment pool, calculated as Dt/(Dd+Dt). (C) Maximum PSII quantum yield and NPQ capacity. (D) Oxygen evolution rates at different light intensities. Errors bars represent SEMs. To avoid the variant light exposure of cells caused by different culture densities, all of the cell lines were harvested at the same culture density (around 3 × 106 cells per milliliter). Significance was determined by two-sided Student’s t tests. P values of significance are indicated (*P < 0.05; **P < 0.01).
The xanthophyll cycle pigments are located in different domains of the thylakoid membranes (25). Detailed investigation of the respective Dd+Dt distribution by analyses of separated pigment–protein complexes demonstrates that the additional Dd+Dt pigments identified in bgs_c (Fig. 3A) are present in the fraction containing the peripheral antennae [fucoxanthin chlorophyll binding protein complexes (FCPs)] and in the free pigment fraction, but not in those of the photosystems (Fig. S7B). Interestingly, the bgs_c mutant also contains a higher number of FCPs per photosystem than the WT cells (Fig. S7C), confirming, on an absolute level, an elevated Dd+Dt amount in the peripheral antenna. The higher amount of FCPs is also reflected by an increased cellular content of fucoxanthin and chlorophyll c (Table S1).
The bgs_b and bgs_c strains furthermore show maximum oxygen evolution rates that are 1.5-fold and 2.2-fold lower compared with WT cells (Fig. 3D). The reduced oxygen evolution rates, PSII quantum yields, and growth rates, as well as the enhanced NPQ capacities and higher de-epoxidation states, reveal that in the bgs mutant cells, less absorbed light energy is effectively transferred from the antennae to the photosystems and less biomass is generated by the same amount of incident light energy compared with the WT cells.
Reduced PtBGS Expression Alters the Thylakoid Membrane Organization.
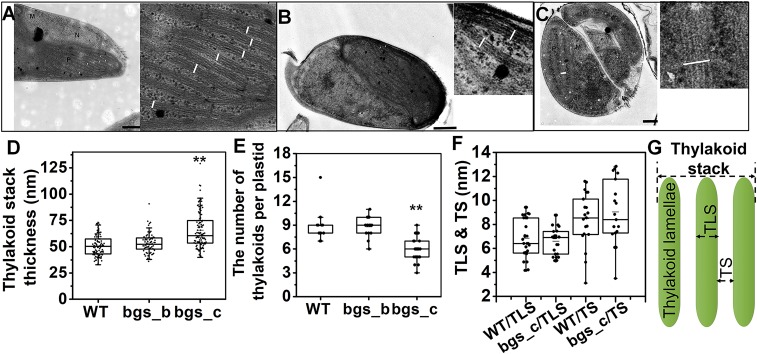
Electron microscopic analyses indicate that reduced PtBGS expression also causes changes in the thylakoid arrangement patterns (Fig. 4 A–E and Fig. S8). The thylakoid stacks of bgs_c exhibit a width of around 66 nm, being significantly enlarged (1.32-fold) compared with the thylakoid thickness of the WT cells (around 50 nm), whereas no significant difference is detectable in bgs_b (Fig. 4D). Interestingly, the increase in thylakoid diameter of bgs_c is accompanied by a decreased (1.37-fold) average number of thylakoid stacks (Fig. 4E). However, the strict triple lamellar morphology of the thylakoid stacks, which is typical for diatoms (26), is unexpectedly altered in bgs_c. Instead of the regular three-lamellae thylakoid stacks, we often observed in the mutant five or even more lamellae forming one thylakoid stack (Fig. 4C, Fig. S8C, and Table S2).
Fig. 4.
Analysis of the thylakoid membrane arrangement in WT and Ptbgs mutants. (A) Cell and thylakoids of WT. (Scale bar: 500 nm.) (B) Cell and thylakoids of bgs_b. (Scale bar: 500 nm.) (C) Cell and thylakoids of bgs_c. (Scale bar: 250 nm.) Lamellae are labeled with white lines. Electron micrographs of WT, bgs_b, and bgs_c were investigated for thickness of the thylakoid stacks (D) and for the number of thylakoid stacks in one plastid (E). (F) Thylakoid luminal space (TLS) and interthylakoidal space (TS) were additionally determined in WT and bgs_c mutant cells. (G) Schematic presentation of the typical thylakoid stack of diatoms. Significance was determined by two-sided Student’s t tests. P values are indicated (**P < 0.01).
Discussion
The Molecular Pathway of Chrysolaminarin Synthesis in Diatoms.
UDPG represents the activated precursor for chrysolaminarin synthesis (9). Two putative UDPG pyrophosphorylase gene models have been described earlier (UGP1 and UGP2) in P. tricornutum (27). According to TargetP and SignalP predictions, UGP1 and UGP2 are targeted to the cytosol and the plastid, respectively. There is experimental evidence that UGP1 is involved in the synthesis of chrysolaminarin, since knockdown of UGP1 significantly reduces the amount of chrysolaminarin in P. tricornutum (28). Here, we experimentally confirm the gene PtBGS (JGI protein ID 56808) to be the chrysolaminarin synthase in P. tricornutum. The predicted structure (Fig. S2) and the localization in the tonoplast (Fig. 1) suggest that PtBGS recruits cytosolic UDPG (likely produced by UGP1) for glucose transfer and mediates translocation of the generated polysaccharide into the vacuole. For synthesis of mature chrysolaminarin, the β-1,3-glucan backbone has to be modified by β-1,6-transglycosylases (TGSs). Two of the three putative TGSs that recently have been identified in P. tricornutum were confirmed to act as β-1,6-TGSs and to be associated with the tonoplast (29), indicating that both synthesis and modification of the β-1,3-glucan backbone may occur at the vacuolar membrane.
Vacuolar β-1,3-Glucan Synthesis Is Embedded in a Complex Metabolic Network.
Arabidopsis thaliana impaired in starch granule initiation (ss3/ss4) shows massively increased amounts of the starch precursor ADP glucose (∼170-fold higher than the WT), as well as the accumulation of soluble sugars (15). Here, we show that in P. tricornutum, reduced chrysolaminarin synthesis leads to an accumulation of soluble sugars; however, there is no considerable accumulation of the respective precursor UDPG (Fig. S5A). It is likely that the residual BGS activity in the silencing mutants may consume some of the available UDPG. In P. tricornutum mutants with a reduced or deleted UGP1 (28, 30), carbohydrate precursors are (at least partially) shunted into lipid synthesis. Our data, together with recent studies on UDP-glucose pyrophosphorylase (ugp) mutants (28, 30), suggest that diatoms may also shuttle precursors into alternative pathways, which reduces the extent of their accumulation. Similarly, UDPG in diatoms might enter the synthesis pathways of, for example, oligosaccharides, cell wall polysaccharides, chitin fibers, or glycoproteins (31). In addition, tight feedback regulation of UGP1 has to be considered as a possible principle to prevent UDPG accumulation. This regulation might result in an increase in phosphorylated precursors and soluble sugars. In fact, we were able to identify slight, but significant, accumulation of specific soluble sugars in the bgs_c line (Fig. S5 B and C). Moreover, the stimulation of lipid biosynthesis (Fig. 2D) can be taken as a further indication for elevated availability of carbohydrates (phosphorylated precursors and soluble sugars).
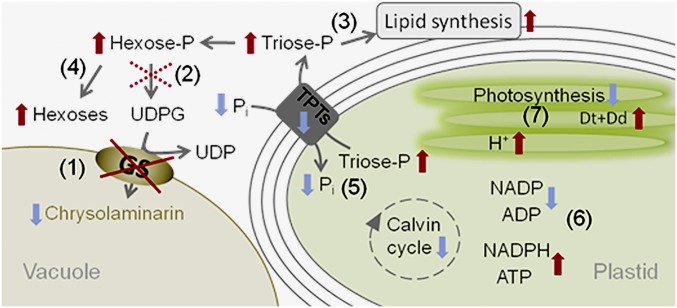
In the following, we propose a possible scenario explaining the observations in the Ptbgs silencing mutants (Fig. 5). Because of reduced chrysolaminarin synthesis, the consumption of phosphorylated sugars, and consequently the delivery of phosphate as well, considerably decreases in the mutant. This may stimulate alternative biosynthetic pathways. Moreover, in land plants, an elevated cytosolic triosephosphate/phosphate ratio decelerates plastidial triosephosphate export via the triosephosphate/phosphate translocator (32), which recently has been identified in different membranes surrounding the complex plastid of P. tricornutum (33). The rising triosephosphate concentrations in the stroma (due to limited export) may lower the activity of the Calvin cycle (feedback regulation), which is an important ATP- and NADPH-demanding pathway (Fig. 5). Indeed, in Chlamydomonas reinhardtii, the rates of NADPH reoxidation in a starchless mutant are attenuated (16). Also, in A. thaliana, the deletion of starch synthesis leads to higher NADPH/NADP ratios (34). Accordingly, the missing turnover of ATP and NADPH restricts the photosynthetic capacity of the mutant (Fig. 3 C and D) due to substrate limitation. This finally leads to an overreduction of the electron transport chain, probably enhancing the thylakoidal proton gradient, thus resulting in the observed de-epoxidation of Dd even under low light intensities (Fig. 3B). As a consequence of the overreduction of the electron transport chain, the higher reduction state of the plastoquinone pool triggers the increase in the total amount of Dd+Dt (35) (Fig. 3A). The additional Dd+Dt pigments are mainly bound to the FCPs (Fig. S7), and thus may contribute to the higher NPQ capacity of the mutants (Fig. 3C).
Fig. 5.
Proposed scenario explaining the observed effects of reduced PtBGS activity in P. tricornutum. Impaired BGS activity results in decreased chrysolaminarin production and lower consumption of UDPG (1). Feedback regulation (dotted cross) of UDPG pyrophosphorylase (2) causes accumulation of hexosephosphates (Hexose-P) and triosephosphates (Triose-P). Phosphorylated sugars can enter alternative pathways (3) and are also dephosphorylated via phosphatases (4), which at least partially lowers the extent of their accumulation and of phosphate (Pi) deficiency. The rising triosephosphate-to-Pi ratio in the cytosol reduces the delivery of triosephosphate from the plastid stroma via triosephosphate/phosphate translocators (TPTs) located in the different plastid membranes. The reduced triosephosphate export and the correspondingly reduced Pi uptake elevate the triosephosphate-to-phosphate ratio in the plastid stroma (5). This situation inhibits key enzymes of the Calvin cycle, and consequently lowers NADPH and ATP consumption (level of NADPH and ATP increases, while that of NADP and ADP drops) (6). NADP reduction and ATP synthesis slow down due to substrate limitation, which reduces the photosynthetic capacity, causes overreduction of the electron transport chain, and increases the proton gradient (H+) at the thylakoid membrane (7). The situation at the thylakoid membrane resembles that of high light conditions and induces corresponding responses, such as an increase in the total Dd+Dt pool and changes in the thylakoid architecture. Changes in metabolism due to reduced PtBGS activity are indicated by arrows (increases are marked in red, decreases are marked in blue).
Interestingly, the resulting growth retardation was not observed under short day conditions (Fig. S6). This indicates that the chrysolaminarin synthesis capacity is only relevant when the metabolic turnover of photosynthetically produced carbohydrates in the cell is lower than the concomitant carbon fixation by photosynthesis, which, under our low light conditions, only occurred during continuous illumination. In line with this, in T. pseudonana, a considerable phenotype related to chrysolaminarin reduction was only observed under silicon limitation (12). In starch-free mutants of A. thaliana, comparable effects on photosynthesis and growth have been recorded only under prolonged high light exposure (36). Our results demonstrate the dependency of diatom photosynthesis on downstream metabolic processes occurring outside of the plastid, but they also highlight the fine-tuning of diatom NPQ capacities in response not only to external stimuli but also to metabolic imbalances.
Reduced β-1,3-Glucan Synthesis Strikingly Alters the Morphological Architecture of the Thylakoids.
In diatoms, high light may lead to a reduction in the number of the thylakoid stacks per plastid, while the number of lamellae per stack remains constant (37). Bgs_c shows a reduction in the thylakoid stack number already under low light conditions (Fig. 4E), and therefore phenotypically resembles diatoms exposed to high light conditions. Strikingly, at the same time, the number of lamellae per stack is increased (Fig. 4C, Fig. S8C, and Table S2). To our knowledge, such a phenotype of P. tricornutum has, to date, only been documented for iron-starved cells (38) and, more pronounced, in cells exposed for a long term to red light (39). Interestingly, in A. thaliana, deletion of the ADP-glucose pyrophosphorylase (adg), inhibiting starch synthesis, leads to hyperstacking of grana thylakoids (34). The number of lamellae per stack is assumed to increase the self-shading of the pigments. This, together with the reduced amount of thylakoids, would result in a decrease of photon absorption, thus partially relieving the reduction pressure in the plastidic electron transport chain in the bgs_c mutant. Strikingly, a typical feature of iron limitation in diatoms is, besides the increase of lamellae per thylakoid stack (38) and the increase of photoprotection capacity (40), a decoupling of antennae from the photosystems, resulting in a lower Fv/Fm value (41). Indeed, the Ptbgs mutants showed a lower Fv/Fm value, a higher photoprotection capacity, more lamellae per stack, and more antennae per photosystem, together with decreased photosynthesis rates. This indicates that some of the antennae might not be functionally connected to the photosystems. Hence, in several aspects, the low chrysolaminarin-dependent syndrome resembles aspects of iron starvation and points to a general stress phenotype induced by the reduced ability to store reduced carbon outside of the plastid.
In summary, by localizing the PtBGS at the tonoplast of P. tricornutum, we confirm that chrysolaminarin synthesis occurs outside of the plastids. Assuming the same functionality of all glucan synthases, and based on its tonoplast localization, the diatom BGS essentially produces a polymeric carbohydrate by secreting a β-glucan chain into the vacuole. There is, however, no evidence that this polymer might form crystalline structures as known, for example, for paramylon in euglenoids. We also reveal that disrupting the cellular ability to produce storage carbohydrates leads to a complex syndrome affecting metabolism, plastid homeostasis, and thylakoid structure. It remains to be solved how the β-glucan strands are arranged structurally within the vacuole, why they do not form a crystalline structure, and how the structure of chrysolaminarin may affect carbohydrate storage and degradation.
Materials and Methods
Cultivation of P. tricornutum.
P. tricornutum (strain UTEX646) was grown at 20 °C in modified f/2 medium with artificial half-concentrated sea salts (16.6 g⋅L−1) and 0.09 μM MnCl2 (42). Cells were grown under continuous white light at 70 μmol photons m−2·s−1 and harvested at the exponential growth phase.
Generation of Vectors for Gene Silencing and eGFP Fusion Genes.
The 110-bp PtBGS antisense fragment flanked by restriction enzymes was generated by Eurofins MWG Operon (SI Materials and Methods). The first intron of Ptntt1 (JGI_ID_49533; 128 bp) was used as a linker between sense and antisense strands. For generation of the silencing plasmid, the 110-bp sense strand, linker, and 110-bp antisense strand were sequentially assembled with the vector pPha-NR [GenBank accession no. JN180663.1 (43)] via standard cloning procedures and a Gibson Assembly Master Mix kit (New England Biolabs GmbH) (44, 45).
Isolation of P. tricornutum genomic DNA and cDNA was performed as described previously (46). For generation of the plasmid pPTV_PtBGS::eGFP, the PtBGS sequence was assembled with the modified pPha-T1 vector (GenBank accession no. AF219942.1), equipped with the egfp gene, using the Gibson Assembly Master Mix kit and standard cloning procedures (44, 45, 47). Vectors were transformed into P. tricornutum using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad) (47).
Analysis of the PtBGS Expression.
Proteins were extracted as described in SI Materials and Methods. A specific antibody against GFP (Invitrogen) and a custom-made antiserum against PtBGS (Agrisera) were used for detection of the fusion protein PtBGS::GFP in the WT and in PtBGS::GFP-expressed cell lines. The expression level of PtBGS in WT and bgs was detected using the antiserum against PtBGS.
Fluorescence Microscopy, Electron Microscopy, and the Staining of Vacuoles in P. tricornutum.
Microscopy was performed as described in SI Materials and Methods. The pattern of vacuoles in P. tricornutum was detected using a green fluorescent vacuolar membrane marker MDY-64 as described by Huang et al. (29) and documented with an Olympus BX51 epifluorescence microscope (Olympus Europe).
Oxygen Evolution and Chlorophyll Fluorescence Measurements.
Oxygen measurements were performed using a Clark-type electrode (Hansatech Instruments Ltd.). Cell densities were adjusted to 5 × 106 cells per milliliter using f/2 medium. Chlorophyll fluorescence was measured using an Aquapen AP-100 (Photon Systems Instruments). The cell densities were adjusted to 4 × 106 cells per milliliter, and cells were all harvested at equal density (roughly 3 × 106 cells per milliliter). The maximum photosynthetic efficiency of PSII was calculated as (Fm − Fo)/Fm = Fv/Fm. Fm and Fo refer to maximum and minimum fluorescence values, respectively, in a dark-adapted state. NPQ was induced by actinic illumination with 600 μmol photons m−2·s−1 and calculated as (Fm − Fm′)/Fm′ (48). Fm and Fm′ were obtained during application of a saturating light flash (2,100 μmol photons m−2·s−1).
Pigment Analysis and Preparation of Pigment–Protein Complexes.
Pigment extraction from cultures that were harvested at equal density (roughly 3 × 106 cells per milliliter) and HPLC analysis were performed as described previously (35, 49). Thylakoids and pigment protein complexes were isolated as described by Lepetit et al. (50). A ratio of β-dodecyl maltoside per chlorophyll a of 30 was used to solubilize thylakoids. Fractions were harvested with a syringe from the bottom to the top to prevent mixture, and absorbance of individual fractions was measured in a GE Healthcare Ultrospec photometer.
Quantification of Chrysolaminarin.
Extraction and colorimetric determination of the glucan were performed according to the methods of Zhu et al. (28) and Granum and Myklestad (51) with slight modifications (SI Materials and Methods).
Quantification of TAGs.
To determine the lipid content in WT, bgs_b, and bgs_c, flow cytometry was applied with a Cell Lab Quanta SC MPL flow cytometer (Beckman Coulter), which is equipped with a 488-nm laser and variable emission filter sets. BODIPY 493/503 (final concentration of 50 ng/mL; Molecular Probes), a lipophilic bright green fluorescent dye (52), was used to monitor the relative TAG accumulation in different cell lines.
Supplementary Material
Acknowledgments
We thank Dr. Heiko Wagner (University of Leipzig) for help with the experiments. We also thank the Electron Microscopy Center, the Bioimaging Center, and the Nanostructure laboratory of the University of Konstanz. We thank Doris Ballert for help in transformation of P. tricornutum cells. We thank Prof. Dr. Wolfgang R. Hess for the support in constructing the phylogenetic tree. This work was supported by a stipend from the China Scholarship Council (Grant 201204910191 to W.H.), by the Zukunftskolleg of the University of Konstanz and the Deutsche Forschungsgemeinschaft (Grant LE 3358/3-1 to B.L.), by the Graduate School Chemical Biology, and by the University of Konstanz.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719274115/-/DCSupplemental.
References
- 1.Falkowski PG, Barber RT, Smetacek V. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;281:200–207. doi: 10.1126/science.281.5374.200. [DOI] [PubMed] [Google Scholar]
- 2.Beattie A, Hirst EL, Percival E. Studies on the metabolism of the Chrysophyceae. Comparative structural investigations on leucosin (chrysolaminarin) separated from diatoms and laminarin from the brown algae. Biochem J. 1961;79:531–537. doi: 10.1042/bj0790531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ford CW, Percival E. Carbohydrates of Phaeodactylum tricornutum. Part I. Preliminary examination of the organism, and characterization of low molecular weight material and of a glucan. J Chem Soc. 1965:7035–7041. [Google Scholar]
- 4.Myklestad S. Production of carbohydrates by marine planktonic diatoms. I. Comparison of nine different species in culture. J Exp Mar Biol Ecol. 1974;15:261–274. [Google Scholar]
- 5.Chiovitti A, et al. The glucans extracted with warm water from diatoms are mainly derived from intracellular chrysolaminaran and not extracellular polysaccharides. Eur J Phycol. 2004;39:117–128. [Google Scholar]
- 6.Michel G, Tonon T, Scornet D, Cock JM, Kloareg B. Central and storage carbon metabolism of the brown alga Ectocarpus siliculosus: Insights into the origin and evolution of storage carbohydrates in eukaryotes. New Phytol. 2010;188:67–81. doi: 10.1111/j.1469-8137.2010.03345.x. [DOI] [PubMed] [Google Scholar]
- 7.Levy A, Epel BL. Cytology of the (1-3)-β-glucan (Callose) in plasmodesmata and sieve plate pores. In: Bacic A, Fincher GB, Stone BA, editors. Chemistry, Biochemistry, and Biology of 1-3 Beta Glucans and Related Polysaccharides. Academic; San Diego: 2009. pp. 439–463. [Google Scholar]
- 8.Stanisich VA, Stone BA. Functional roles of (1,3)-β-glucans and related polysaccharides: Prokaryotes. In: Bacic A, Fincher GB, Stone BA, editors. Chemistry, Biochemistry, and Biology of 1-3 Beta Glucans and Related Polysaccharides. Academic; San Diego: 2009. pp. 327–352. [Google Scholar]
- 9.Roessler PG. UDPglucose pyrophosphorylase activity in the diatom Cyclotella cryptica. Pathway of chrysolaminarin synthesis. J Phycol. 1987;23:494–498. [Google Scholar]
- 10.Bowler C, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 11.Chauton MS, Winge P, Brembu T, Vadstein O, Bones AM. Gene regulation of carbon fixation, storage, and utilization in the diatom Phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiol. 2013;161:1034–1048. doi: 10.1104/pp.112.206177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hildebrand M, et al. Effects of chrysolaminarin synthase knockdown in the diatom Thalassiosira pseudonana: Implications of reduced carbohydrate storage relative to green algae. Algal Res. 2017;23:66–77. [Google Scholar]
- 13.Suzuki E, Suzuki R. Variation of storage polysaccharides in phototrophic microorganisms. J Appl Glycosci. 2013;60:21–27. [Google Scholar]
- 14.Tanaka Y, et al. Glucan synthase-like 2 is indispensable for paramylon synthesis in Euglena gracilis. FEBS Lett. 2017;591:1360–1370. doi: 10.1002/1873-3468.12659. [DOI] [PubMed] [Google Scholar]
- 15.Ragel P, et al. Loss of starch granule initiation has a deleterious effect on the growth of Arabidopsis plants due to an accumulation of ADP-glucose. Plant Physiol. 2013;163:75–85. doi: 10.1104/pp.113.223420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan A, et al. Metabolic and photosynthetic consequences of blocking starch biosynthesis in the green alga Chlamydomonas reinhardtii sta6 mutant. Plant J. 2015;81:947–960. doi: 10.1111/tpj.12783. [DOI] [PubMed] [Google Scholar]
- 17.Verma DP, Hong Z. Plant callose synthase complexes. Plant Mol Biol. 2001;47:693–701. doi: 10.1023/a:1013679111111. [DOI] [PubMed] [Google Scholar]
- 18.Okada H, et al. Multiple functional domains of the yeast l,3-beta-glucan synthase subunit Fks1p revealed by quantitative phenotypic analysis of temperature-sensitive mutants. Genetics. 2010;184:1013–1024. doi: 10.1534/genetics.109.109892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz-Herrera J, Ortiz-Castellanos L. Analysis of the phylogenetic relationships and evolution of the cell walls from yeasts and fungi. FEMS Yeast Res. 2010;10:225–243. doi: 10.1111/j.1567-1364.2009.00589.x. [DOI] [PubMed] [Google Scholar]
- 20.Trentacoste EM, et al. Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc Natl Acad Sci USA. 2013;110:19748–19753. doi: 10.1073/pnas.1309299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levitan O, Dinamarca J, Zelzion E, Gorbunov MY, Falkowski PG. An RNA interference knock-down of nitrate reductase enhances lipid biosynthesis in the diatom Phaeodactylum tricornutum. Plant J. 2015;84:963–973. doi: 10.1111/tpj.13052. [DOI] [PubMed] [Google Scholar]
- 22.Smith SR, et al. Transcript level coordination of carbon pathways during silicon starvation-induced lipid accumulation in the diatom Thalassiosira pseudonana. New Phytol. 2016;210:890–904. doi: 10.1111/nph.13843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stransky H, Hager A. The carotenoid pattern and the occurrence of the light induced xanthophyll cycle in various classes of algae. Arch Mikrobiol. 1970;71:164–190. [PubMed] [Google Scholar]
- 24.Goss R, Lepetit B. Biodiversity of NPQ. J Plant Physiol. 2015;172:13–32. doi: 10.1016/j.jplph.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Lepetit B, Volke D, Gilbert M, Wilhelm C, Goss R. Evidence for the existence of one antenna-associated, lipid-dissolved and two protein-bound pools of diadinoxanthin cycle pigments in diatoms. Plant Physiol. 2010;154:1905–1920. doi: 10.1104/pp.110.166454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pyszniak AM, Gibbs S. Immunocytochemical localization of photosystem I and the fucoxanthin-chlorophylla/c light-harvesting complex in the diatom Phaeodactylum tricornutum. Protoplasma. 1992;166:208–217. [Google Scholar]
- 27.Kroth PG, et al. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS One. 2008;3:e1426. doi: 10.1371/journal.pone.0001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu BH, et al. Silencing UDP-glucose pyrophosphorylase gene in Phaeodactylum tricornutum affects carbon allocation. N Biotechnol. 2016;33:237–244. doi: 10.1016/j.nbt.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Huang W, Río Bártulos C, Kroth PG. Diatom vacuolar 1,6-beta-transglycosylases can functionally complement the respective yeast mutants. J Eukaryot Microbiol. 2016;63:536–546. doi: 10.1111/jeu.12298. [DOI] [PubMed] [Google Scholar]
- 30.Daboussi F, et al. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat Commun. 2014;5:3831. doi: 10.1038/ncomms4831. [DOI] [PubMed] [Google Scholar]
- 31.Gügi B, et al. Diatom-specific oligosaccharide and polysaccharide structures help to unravel biosynthetic capabilities in diatoms. Mar Drugs. 2015;13:5993–6018. doi: 10.3390/md13095993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flügge U-I. Phosphate translocation in the regulation of photosynthesis. J Exp Bot. 1995;46:1317–1323. [Google Scholar]
- 33.Moog D, Rensing SA, Archibald JM, Maier UG, Ullrich KK. Localization and evolution of putative triose phosphate translocators in the diatom Phaeodactylum tricornutum. Genome Biol Evol. 2015;7:2955–2969. doi: 10.1093/gbe/evv190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Häusler RE, et al. Chlororespiration and grana hyperstacking: How an Arabidopsis double mutant can survive despite defects in starch biosynthesis and daily carbon export from chloroplasts. Plant Physiol. 2009;149:515–533. doi: 10.1104/pp.108.128124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lepetit B, et al. High light acclimation in the secondary plastids containing diatom Phaeodactylum tricornutum is triggered by the redox state of the plastoquinone pool. Plant Physiol. 2013;161:853–865. doi: 10.1104/pp.112.207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitz J, et al. Defects in leaf carbohydrate metabolism compromise acclimation to high light and lead to a high chlorophyll fluorescence phenotype in Arabidopsis thaliana. BMC Plant Biol. 2012;12:8. doi: 10.1186/1471-2229-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janssen M, Bathke L, Marquardt J, Krumbein WE, Rhiel E. Changes in the photosynthetic apparatus of diatoms in response to low and high light intensities. Int Microbiol. 2001;4:27–33. doi: 10.1007/s101230100005. [DOI] [PubMed] [Google Scholar]
- 38.Allen AE, et al. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc Natl Acad Sci USA. 2008;105:10438–10443. doi: 10.1073/pnas.0711370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bína D, Herbstová M, Gardian Z, Vácha F, Litvín R. Novel structural aspect of the diatom thylakoid membrane: Lateral segregation of photosystem I under red-enhanced illumination. Sci Rep. 2016;6:25583. doi: 10.1038/srep25583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taddei L, et al. Multisignal control of expression of the LHCX protein family in the marine diatom Phaeodactylum tricornutum. J Exp Bot. 2016;67:3939–3951. doi: 10.1093/jxb/erw198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behrenfeld MJ, Milligan AJ. Photophysiological expressions of iron stress in phytoplankton. Annu Rev Mar Sci. 2013;5:217–246. doi: 10.1146/annurev-marine-121211-172356. [DOI] [PubMed] [Google Scholar]
- 42.Guillard RRL. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH, editors. Culture of Marine Invertebrate Animals. Plenum Press; New York: 1975. pp. 29–60. [Google Scholar]
- 43.Stork S, et al. Distribution of the SELMA translocon in secondary plastids of red algal origin and predicted uncoupling of ubiquitin-dependent translocation from degradation. Eukaryot Cell. 2012;11:1472–1481. doi: 10.1128/EC.00183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, et al. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 45.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 46.Haimovich-Dayan M, et al. The role of C4 metabolism in the marine diatom Phaeodactylum tricornutum. New Phytol. 2013;197:177–185. doi: 10.1111/j.1469-8137.2012.04375.x. [DOI] [PubMed] [Google Scholar]
- 47.Zaslavskaia LA, et al. Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J Phycol. 2000;36:379–386. [Google Scholar]
- 48.Bilger W, Björkman O. Role of the xanthophyll cycle in photoprotection elucidated by measurements of light-induced absorbance changes, fluorescence and photosynthesis in leaves of Hedera canariensis. Photosynth Res. 1990;25:173–185. doi: 10.1007/BF00033159. [DOI] [PubMed] [Google Scholar]
- 49.Jakob T, et al. Activation of diadinoxanthin de-epoxidase due to a chlororespiratory proton gradient in the dark in the diatom Phaeodactylum tricornutum. Plant Biol. 1999;1:76–82. [Google Scholar]
- 50.Lepetit B, et al. Spectroscopic and molecular characterization of the oligomeric antenna of the diatom Phaeodactylum tricornutum. Biochemistry. 2007;46:9813–9822. doi: 10.1021/bi7008344. [DOI] [PubMed] [Google Scholar]
- 51.Granum E, Myklestad SM. A simple combined method for determination of β-1,3-glucan and cell wall polysaccharides in diatoms. Hydrobiologia. 2002;477:155–161. [Google Scholar]
- 52.Govender T, Ramanna L, Rawat I, Bux F. BODIPY staining, an alternative to the Nile Red fluorescence method for the evaluation of intracellular lipids in microalgae. Bioresour Technol. 2012;114:507–511. doi: 10.1016/j.biortech.2012.03.024. [DOI] [PubMed] [Google Scholar]