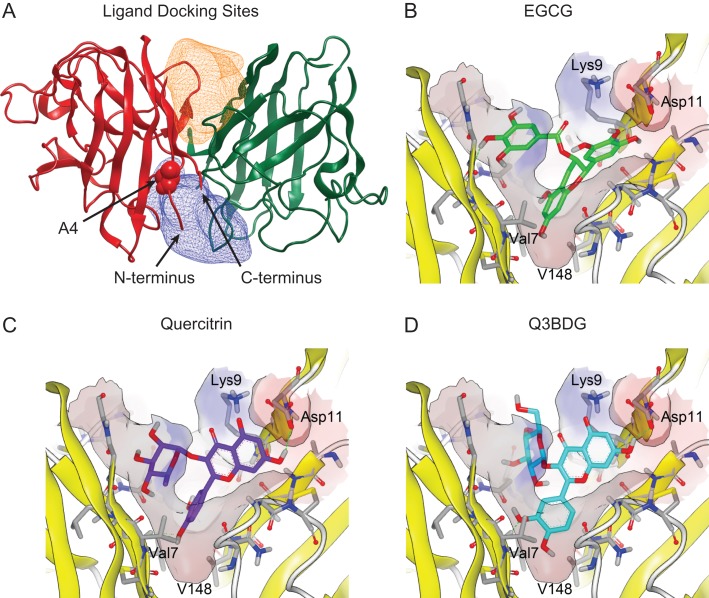
Fig. 1.
Computer docking of drugs into the SOD1 structure. The structure of wild-type SOD1 (PDB ID: 1SPD) is shown with the simulated regions highlighted. (A) The constructed binding pockets are displayed as meshes that encompass the volume that drugs were docked into. ‘Pocket 1’ is on top of the depicted dimer interface while ‘Pocket 2’ is on the bottom. These two regions around the dimer interface were isolated from this structure using the OpenEye GUI program ‘make_receptor’. The alanine at residue four for the subunit on the left is shown as space-filling balls and labeled. The N- and C-termini of this left subunit are indicated with arrows. (B–D) Small molecules from the SWEETLEAD database were allowed to dock into these regions using OpenEye FRED, and their binding energies were evaluated using Chemgauss4 scoring. The predicted binding of EGCG, quercitrin, and Q3BDG to Pocket 1 are shown in a similar orientation as the overall SOD1 molecule depicted in panel (A). Structures were rendered using VIDA 4.3.0.4. The predicted binding of Nice, BPP, CDCA, and HupA are shown in Supplementary Figure 5.