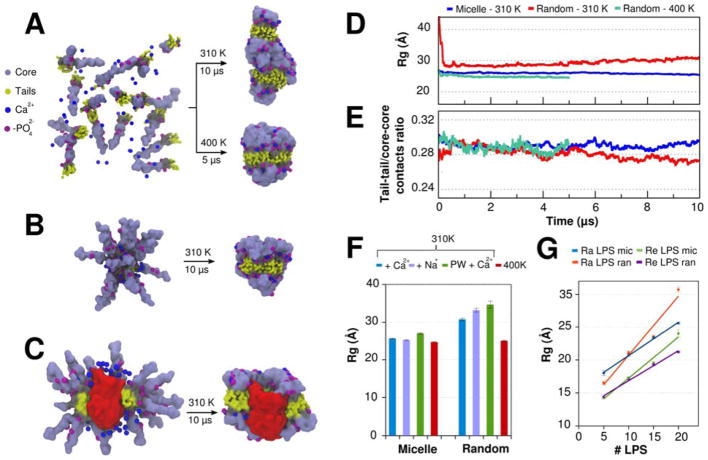
Figure 6.
(A–C) Initial structure and last snapshot of 10-μs MD simulations (at 310 K and 1 bar) performed with different Ra LPS systems. (A) 20 Ra LPS molecules were randomly placed and self-assembled during the simulations. In this case, an auxiliary 5-μs MD simulation was also performed at higher temperature (400 K) and fixed density (NVT ensemble). (B) 20 Ra LPS molecules were preassembled in a micelle configuration. (C) 40 Ra LPS molecules and one monomer of OmpF in a micelle configuration. For sake of clarity, only half of the LPS molecules are shown. (D) Time-series of the radius of gyration (Rg) of 20 Ra LPS systems. (E) Time-series of the tail-tail/core-core contacts ratio of 20 Ra LPS systems. Tail-tail and core-core contacts were defined using a distance cutoff of 6 Å between the beads. (F) Average radius of gyration of 20 Ra LPS molecules in four different conditions: (i) with Ca2+ ions added near to the phosphate groups of lipid A; (ii) with Na+ ions replacing Ca2+ ions; (iii) with Ca2+ ions and polarizable water model; (iv) with Ca2+ ions and higher temperature (400K). (G) Average radius of gyration of 5, 10, 15, and 20 Ra and Re LPS molecules in MD simulations performed with random and micelle initial configurations.