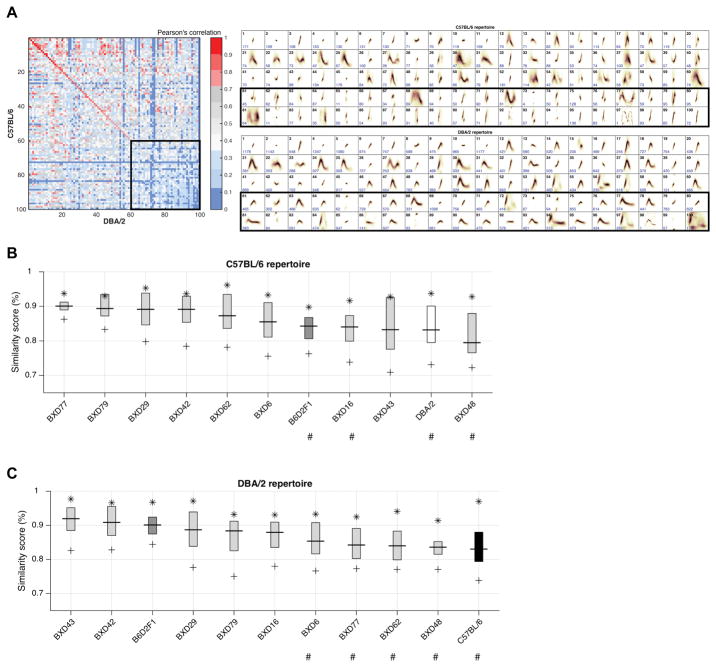
Figure 6. Similarity metrics used to compare repertoire unit type across strains.
(A) Similarity Matrix (left panel) of the spectral types of pairs of repertoire units (RUs) learned from the C57BL/6 and DBA/2 datasets (right panels). The matrix diagonal gives the Pearson correlation for sequential pairs of C57BL/6 and DBA/2 RUs ranked from most to least similar (e.g., Unit 1 in both repertories are highly similar). Comparison of RU types is performed by centering RUs along the time and frequency axes and then by sequentially pairing units of greatest to least shape similarity, independent of frequency of use. The parental strains produce both highly similar (e.g., unit 1–40) and distinct (e.g., units 60–100) repertoire units. Black boxes highlight RUs that show low similarity across the parental strains.
(B–C) The Similarity Boxplots determined by comparing the similarity of RU types as a function of how frequently the RU is used by each of the ‘comparison’ strains (X-axis) in comparison to the C57BL/6 (B) and DBA/2 (C) ‘reference’ strains. The Y-axis is the % Similarity Score (average Pearson correlations) between collections of RUs compared between the reference and comparison repertoires. The star (*) denotes the Pearson correlations for the top 5% of the most frequently used RUs, where the boxplot shows the mean and interquartile range of these correlations, and the plus sign (+) shows the correlation of the top 95% of the most frequency used RUs. # Indicates strains with statistically significant (P < 0.05) differences in mean repertoire similarity compared to the reference repertoire.