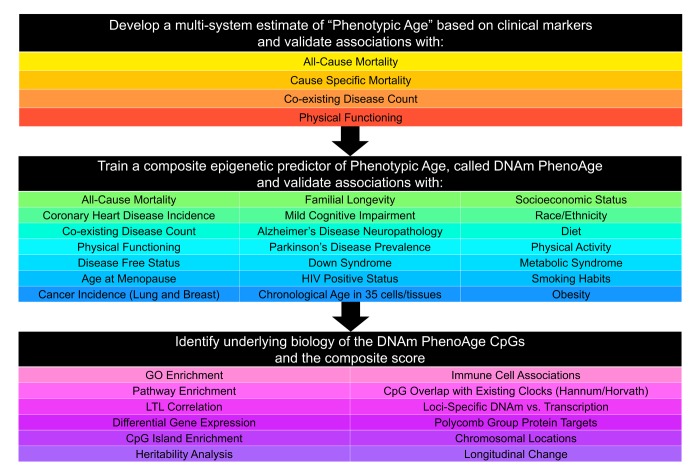
Figure 1.
Roadmap for developing DNAm PhenoAge. The roadmap depicts our analytical procedures. In step 1, we developed an estimate of ‘Phenotypic Age’ based on clinical measure. Phenotypic age was developed using the NHANES III as training data, in which we employed a proportional hazard penalized regression model to narrow 42 biomarkers to 9 biomarkers and chronological age. This measure was then validated in NHANES IV and shown to be a strong predictor of both morbidity and mortality risk. In step 2, we developed an epigenetic biomarker of phenotypic age, which we call DNAm PhenoAge, by regressing phenotypic age (from step 1) on blood DNA methylation data, using the InCHIANTI data. This produced an estimate of DNAm PhenoAge based on 513 CpGs. We then validated our new epigenetic biomarker of aging, DNAm PhenoAge, using multiple cohorts, aging-related outcomes, and tissues/cells. In step 3, we examined the underlying biology of the 513 CpGs and the composite DNAm PhenoAge measure, using a variety of complementary data (gene expression, blood cell counts) and various genome annotation tools including chromatin state analysis and gene ontology enrichment.