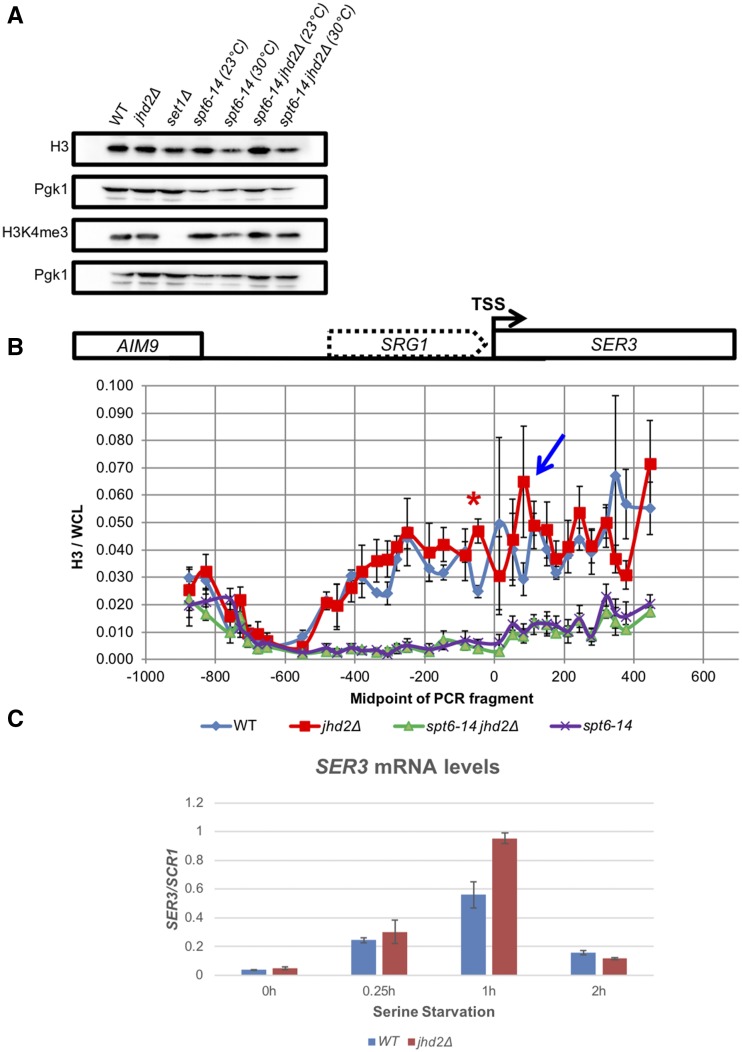
Figure 5.
JHD2 deletion does not rescue Spt6-Spn1 mutants by compensating for defects in chromatin structure. (A) Western blot detection of H3K4me3, pan-H3, and Pgk1 are shown from extracts of wildtype and mutant cells grown at 23°C (permissive) and 30°C (semi-permissive temperature). Pgk1 serves as a loading control. (B) Histone H3 ChIP was performed on strains grown at 30°C (semi-permissive temperature) across the SRG1-SER3 locus (schematic of locus shown above graph). H3 ChIP signal was normalized to signal from whole cell lysate (WCL) DNA. The transcription start site of SER3 is positioned at 0 and for SRG1 at -480. Error bars represent standard error of the mean. Significance as calculated by a two-tailed student’s t-test is shown where *P < 0.05. A blue arrow denotes an additional H3 ChIP peak in jhd2∆ that was not statistically significant. (C) SER3 transcript level was measured by RT-qPCR at specific intervals after serine withdrawal in wildtype and jhd2∆ mutants. SER3 signal was normalized to SCR1.