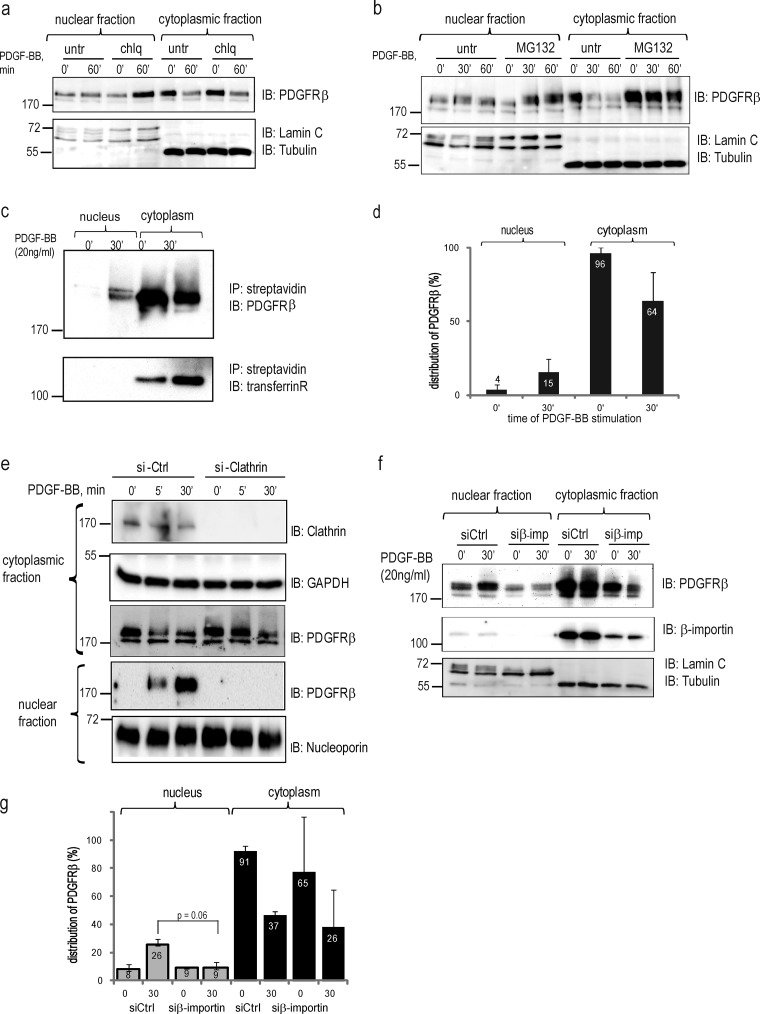
Figure 3.
Characterization of nuclear translocation pathways. (a) PDGFRβ accumulates in the nucleus after inhibition of the endosomal–lysosomal pathway. Nuclear and cytoplasmic fractions treated or not with chloroquine (chlq) were immunoblotted with total PDGFR antibody (top), lamin A/C, and α-tubulin (bottom). A representative experiment of three biological repeats is shown. (b) Accumulation of PDGFRβ in the nucleus upon inhibition of proteasomal degradation. Nuclear and cytoplasmic fractions treated or not with MG132 were immunoblotted with total PDGFRβ antibody (top) or nuclear and cytoplasmic markers (bottom). (c) Biotinylated cell surface receptor is translocated to the nucleus. Cytoplasmic and nuclear fractions were prepared from biotinylated cells stimulated with PDGF-BB. Biotinylated proteins were precipitated with streptavidin agarose and immunoblotted for total PDGFRβ (top) or transferrin receptor (bottom). (d) Quantification of the distribution of biotinylated PDGFRβ was performed as described in Fig. 1 b. Error bars indicate SD. (e) Nuclear translocation of PDGFRβ is inhibited by siRNA knockdown of clathrin. The efficiency of clathrin siRNA knockdown was determined with an anticlathrin antibody (top). The levels of PDGFRβ in the cytoplasmic and nuclear fractions were determined by immunoblotting (middle). The purity of the fractions was confirmed by immunoblotting for GAPDH (a cytoplasmic marker) or lamin A/C. (f) Nuclear translocation of PDGFRβ is decreased upon siRNA knockdown of β-importin. The level of PDGFRβ in the cytoplasmic and nuclear fractions was determined by immunoblotting with total PDGFR antibody (top); the purity of fractionations was confirmed by blotting for lamin A/C and α–tubulin (bottom). The efficiency of the knockdown was determined by immunoblotting with anti–β-importin antibody (middle). (g) Quantification of PDGFRβ distribution upon β-importin siRNA knockdown was performed as described above and was based on three independent experiments. Molecular mass was measured in kilodaltons. Error bars indicate SD. IB, immunoblotting; IP, immunoprecipitation.