Figure 3. Mapping and manipulating CMV’s transcriptional ‘accelerator’ circuit.
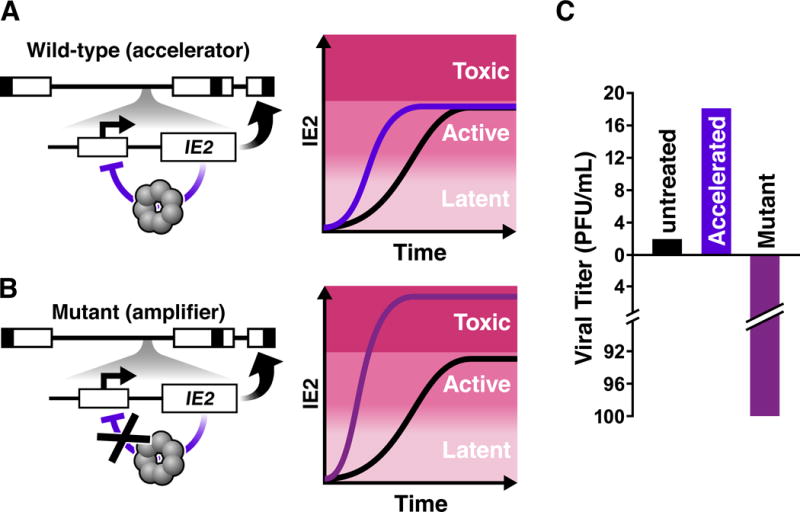
A) (Left) Schematic of the CMV genome (linear form) with the IE2 negative-feedback ‘accelerator’ circuit. As is common, boxes represent terminal and inverted repeat regions of the genome, respectively, while lines represent unique long and unique short genomic regions. The Major Immediate-Early (MIE) locus is encoded within the unique long region of the genome. For simplicity, one the MIE promoter and IE2 are depicted. IE2 is an obligate transactivator of CMV early genes (thick curved arrow). However, IE2 is also cytotoxic at high levels and auto-represses its own MIE promoter as a high-order homo-multimer, forming a highly self-cooperative negative-feedback circuit that limits IE2 expression (purple blunted arrow). (right) Schematic of single-cell imaging trajectories of IE2 protein levels, adapted from (59). This highly cooperative IE2 negative feedback allows accelerated IE2 kinetics but precludes IE2 levels from increasing to levels where IE2 could be cytotoxic (i.e. acts as an ‘accelerator’).
B) (left) Schematic of the CMV recombinant mutant ‘amplifier’ circuit where two nucleotides in the 14 base-pair IE2 DNA-binding site (the ‘cis repression sequence’) within the MIE promoter were mutated. (right) The negative-feedback ‘amplifier’ mutant generates increased IE2 levels, reaching into the cytotoxic regime, upon promoter activation.
C) The IE2 circuit’s effect on viral replicative fitness. Accelerating IE2 accumulation in single-cells by a few hours yields a replicative fitness advantage for the virus (~5–10 fold depending on viral strain). In contrast, disrupting IE2 feedback, by just a two-nucleotide mutation in the MIE promoter, amplifies IE2 levels in individual cells, causing substantial cytotoxicity to the host cell and reducing viral output > 100-fold (Adapted from (59)).
