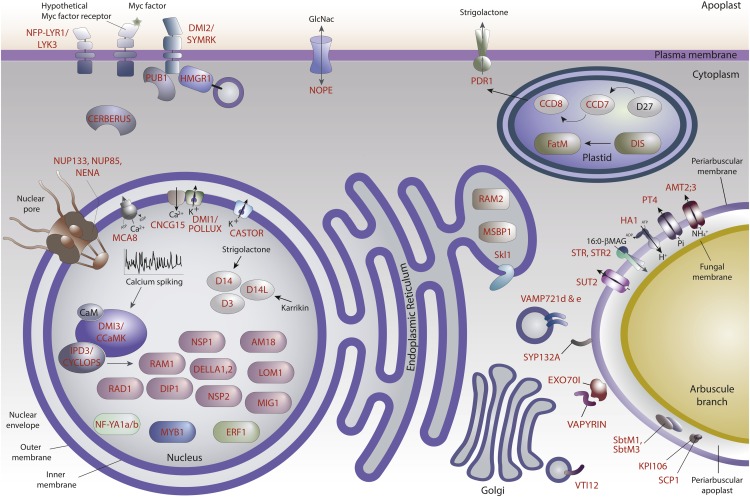
Figure 1.
An Overview of Plant Proteins That Play Key Roles in AM Symbiosis.
Genetic analyses enabled the identification of genes required for AM symbiosis and the mutants show phenotypes that range from an absence of colonization to a quantitative reduction in plant colonization by AM fungi, and in some cases, an increase in the colonization. For many of these proteins, their subcellular location provides additional clues as to their ultimate function during AM symbiosis. For example, receptors embedded in the plasma membrane perceive external signal molecules that trigger calcium spiking in the nucleus. However, some receptors, such as rice D14 and D14L, could be localized in the nucleus. The downstream transcriptional response, mediated by many transcriptional regulators, results in the production of proteins involved in the cellular remodeling and metabolic regulation necessary for symbiosis. This includes enzymes involved in fatty acid production that are localized in plastids and endoplasmic reticulum and transporters localized in the periarbuscular membrane that are involved in the exchange of nutrients with the AM fungus. This figure shows genes for which there is a knockout/knockdown symbiosis phenotype combined with some direct or indirect information about the location of the encoded protein. The gene names used most frequently are indicated in red and also shown in Table 1.