Figure 3.
Lipid Metabolism in the Colonized Cell and Transfer to the AM Fungus
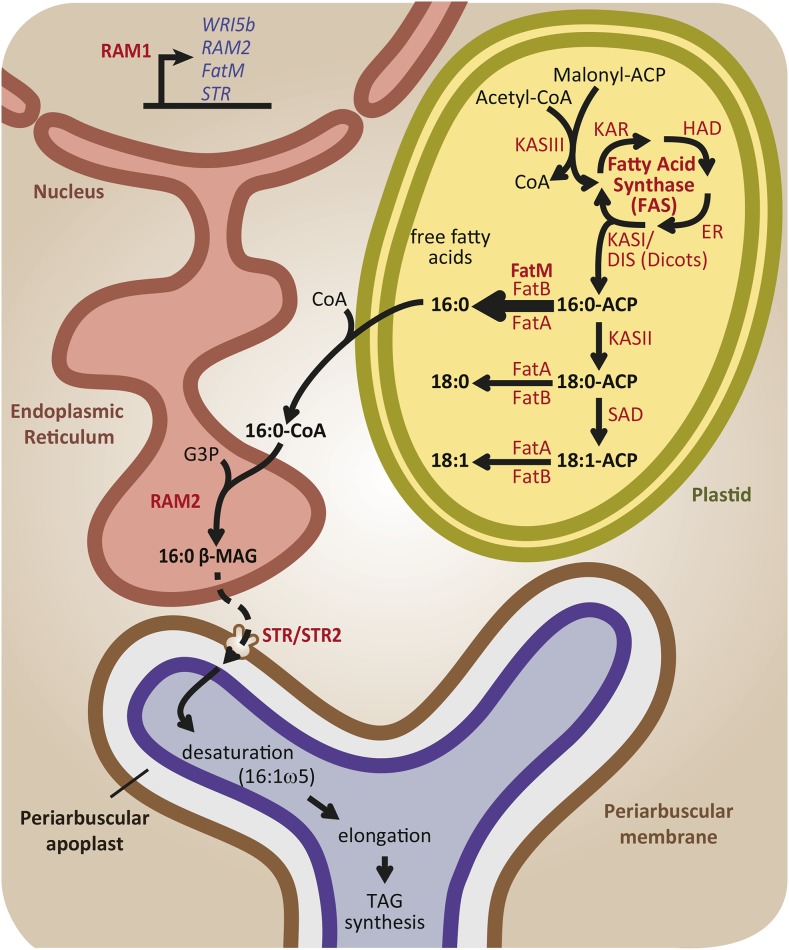
Lipid biosynthesis increases in cortical cells containing arbuscules to produce lipids that will be transferred to the AM fungus. Regulation of lipid biosynthesis begins with the transcriptional induction of several genes encoding enzymes involved in lipid biosynthesis by a transcriptional regulator, RAM1. The downstream targets of RAM1 identified so far are present exclusively in plant species that form AM symbiosis, as is RAM1 itself, and the encoded enzymes regulate key points in fatty acid and monoacylglycerol biosynthesis. Plant de novo fatty acid synthesis begins in plastids with the elongation of the two-carbon acyl moiety of malonyl-ACP. The elongation is performed by the multimeric enzyme fatty acid synthase whose product is a 16-carbon acyl molecule attached to an acyl carrier protein. In dicots and monocots but not in grasses, an additional KASI protein named DIS is responsible for increasing the biosynthesis of fatty acids in plastids of colonized cells. During AM symbiosis, the thioesterase FatM boosts the release of 16:0 fatty acids (palmitic acid) that, when attached to CoA, are used as a substrate by RAM2 to produce 16:0 β-MAG. 16:0 β-MAG or a derivative of this molecule is subsequently exported across the PAM by the half ABC transporters STR and STR2 where it is accessed by the AM fungus. The fungus modifies the 16:0 acyl moiety through desaturation and elongation reactions to produce a variety of fungal lipids. The main proteins (red) and lipid products (black) involved in redirecting lipid metabolism in the colonized cell are shown. Genes regulated by RAM1 are shown in blue.