Figure 4.
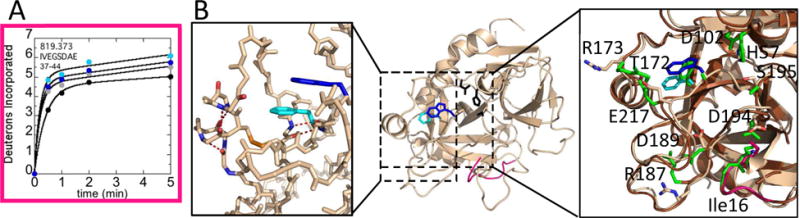
A) Deuterium incorporation over 5 min into residues 16-23CT (residues 36-44; MH+ 819.373) for WT thrombin at 100 mM NaCl (grey) and 300 mM NaCl (black), and for the W215A mutant at 100 mM NaCl (cyan) and 300 mM NaCl (blue). B) Structure of WT thrombin (wheat; PDB 1PPB) highlighting residues 16-23CT (pink). The sidechains of Trp215CT (blue), Phe227CT (cyan), and the catalytic triad (black) are shown as sticks. Accelerated MD simulations identified 5 H-bonds (red-dotted lines) within WT thrombin (left) that broke during simulations of W215A. The transient structure observed during the W215A simulation (brown) overlaying the structure of WT thrombin (right). The side chains of His57CT, Asp102CT, Thr172CT, Arg173CT, Arg 187CT, Asp 189CT, Asp194CT, Ser195CT, Trp215CT (blue), Glu217CT, and Phe227CT (cyan). The 140sCT loop is hidden for clarity. The backbone of Ile16CT (pink) is also shown as sticks. The corresponding side chains in the W215A structure, including Ala215CT, are colored green and are shown as sticks.
