Abstract
Background
Magnetoencephalography (MEG) and Electroencephalography (EEG) are noninvasive techniques to study the electrophysiological activity of the human brain. Thus, they are well suited for real-time monitoring and analysis of neuronal activity. Real-time MEG/EEG data processing allows adjustment of the stimuli to the subject’s responses for optimizing the acquired information especially by providing dynamically changing displays to enable neurofeedback.
New Method
We introduce MNE Scan, an acquisition and real-time analysis software based on the multipurpose software library MNE-CPP. MNE Scan allows the development and application of acquisition and novel real-time processing methods in both research and clinical studies. The MNE Scan development follows a strict software engineering process to enable approvals required for clinical software.
Results
We tested the performance of MNE Scan in several device-independent use cases, including, a clinical epilepsy study, real-time source estimation, and Brain Computer Interface (BCI) application.
Comparison with Existing Method(s)
Compared to existing tools we propose a modular software considering clinical software requirements expected by certification authorities. At the same time the software is extendable and freely accessible.
Conclusion
We conclude that MNE Scan is the first step in creating a device-independent open-source software to facilitate the transition from basic neuroscience research to both applied sciences and clinical applications.
Keywords: MEG/EEG, Neurofeedback/BCI, Data Acquisition, Data Processing, Medical Software
1. Introduction
Magnetoencephalography (MEG) [1, 2] and Electroencephalography (EEG) [3] are widely used to measure the electrophysiological activity of the human brain. During recent years several free software packages such as MNE-C [4], MNE-Python [5], Brainstorm [6], EEGLAB [7], Fieldtrip [8] and OpenMEEG [9] have been created to analyze MEG and EEG data off-line, i.e., after the data collection is complete and the data have been stored. In addition, most clinical studies employ proprietary commercial software tools such as BESA [10], Curry [11], ASA [12], EMSE [13] to ensure compliance with existing national and international regulations for medical software development.
MEG and EEG provide continuous, high-temporal resolution data streams, which enables monitoring brain dynamics in real-time. Therefore, a growing interest in real-time processing of MEG and EEG data exists. For example, it is an indispensable preliminary step for all neurofeedback and Brain Computer Interface (BCI) applications employing MEG/EEG. With real-time processing, relevant features can be extracted and classified immediately from the incoming data stream at the sensor level or source level in the brain. Based on the measured and classified brain state, subsequent devices can be integrated into a closed-loop setup, to present stimuli dependent on the brain state [14]. Furthermore, real-time data analysis allows to evaluate the general measurement setup, analyze the subject’s responses, and in assess the feasibility of the research idea early on to save time and resources, c.f. Bogner et. al. [15]. Real-time MEG/EEG analysis tools can therefore be of great use to both basic researchers and clinicians.
Software packages which provide real-time data processing and analysis can predominantly be found in the fields of neurofeedback and BCI research. Well-known such packages include BCILAB [16], BCI2000 [17], Fieldtrip [8], OpenVibe [18] and OpenBCI [19], all shared under open-source licenses, see Table 1. Investigations of BCI and neurofeedback in clinical studies have been proposed as well. In these studies, either an existing software toolbox was used, or completely new software was proposed. However, in scope of the MEG and EEG community there have been no focused efforts to generalize such software for clinical environments. This might be due to the demanding requirements related to writing software which can obtain medical software clearance, i.e., as defined in the IEC 62304 standard [20]. IEC 62304 harmonizes with certification authorities such as the Food and Drug Administration (FDA) or Conformité Européenne (CE). Importantly, if real-time software has regulatory clearance, it simplifies the transition between clinical settings and in basic research studies, opening new possibilities for cross fertilization. Free-to-use open-source software, which offers a software development framework and at the same time considers regulatory requirements is to our knowledge currently not available. Table 1 presents a comparison between state-of-the-art software packages for real-time data processing.
Table 1.
Comparison of state-of-the-art real-time processing software, including accessibility, dependence on third party software tools, modality support and medical approval.
| MNE Scan | OpenVibe | OpenBCI | Fieldtrip | BCI2000 | BCILAB | |
|---|---|---|---|---|---|---|
| Real-Time capable | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Number of dependencies | 1 | 7 | 1 | 1 | 4 | 1 |
| Open-source access | ✓ | ✓(until Dec 2016) | ✓ | ✓ | ✓ | ✓ |
|
Supported modalities (status 09/2017) |
MEG, EEG | EEG | EEG | MEG, EEG, fMRI | MEG, EEG | EEG |
| Platform independent | ✓(Win, Mac, Linux) | ✓(Win, Linux) | ✓(Win, Mac, Linux) | ✓(Win, Mac, Linux) | ✓(Win, Mac, Linux) | ✓(Win, Mac, Linux) |
|
Free of charge (no add. license required, e.g. Matlab) |
✓ | ✓ | ✓ | |||
| Considers IEC 62304 | ✓ |
Developing basic research analysis methods in a software which meets regulatory requirements could potentially lead to faster integration of these tools into clinical use. When creating new software for clinical studies, risk analysis needs to be conducted and proper software design process must be established. Thus, in case of a failure the consequences can be predicted and the exposure to hazardous situations minimized. Among others this is a prerequisite for FDA, CE and other medical approvals.
Here we introduce MNE Scan [21, 22], a new standalone acquisition and real-time processing application for research and clinical studies. MNE Scan employs MNE-CPP, a software framework based on the MNE software package [4], written in C++. MNE Scan offers device connectors, real-time processing tools, and an easy-to-use Graphical User Interfaces (GUI). MNE Scan makes use of MNE-CPP libraries which allow the processing of electrophysiological data streams and formats, e.g., the established Elekta Neuromag® FIFF file format. MNE Scan can be compiled on the common operating systems, e.g., Windows, Linux, and MacOS and real-time operation systems, such as QNX or VxWorks. Additionally, it can be deployed to smartphones, tablets and embedded systems. The plug-in-based architecture of MNE Scan allows for adding new real-time features for individual use cases. The plug-ins can be connected to form complex processing chains. The results can be visualized in real-time and are accessible during the ongoing measurement session. MNE Scan, like MNE-CPP, is distributed under open source BSD license (clause 3) and freely accessible (https://www.mne-cpp.org).
With MNE Scan we are targeting three main goals:
To provide open-source tools for the analysis during the actual measurement session and thus make results available in real-time.
To follow medical regulatory requirements and thus also enable MNE Scan usage in patient-critical studies, i.e., clinical ones.
To reduce the time between the development of new real-time research methods and the actual application of these methods in basic research and clinical studies.
In this paper, we describe the internal structure of MNE Scan and its real-time processing features as well as give an overview of its development process. Finally, we present results based on different use case scenarios.
2. Software Design and Implementation
2.1. The MNE-CPP Framework
MNE Scan is part of the MNE-CPP project. MNE-CPP is designed as a two-layer framework, see Figure 1. The library layer (Li) provides the core functionalities, which can be used during development and loaded by applications during runtime. The stand-alone applications, examples and tests are realized within the application layer of MNE-CPP. As a stand-alone software, MNE Scan (Sc) is part of the application layer and uses the functionality of the library layer.
Figure 1.
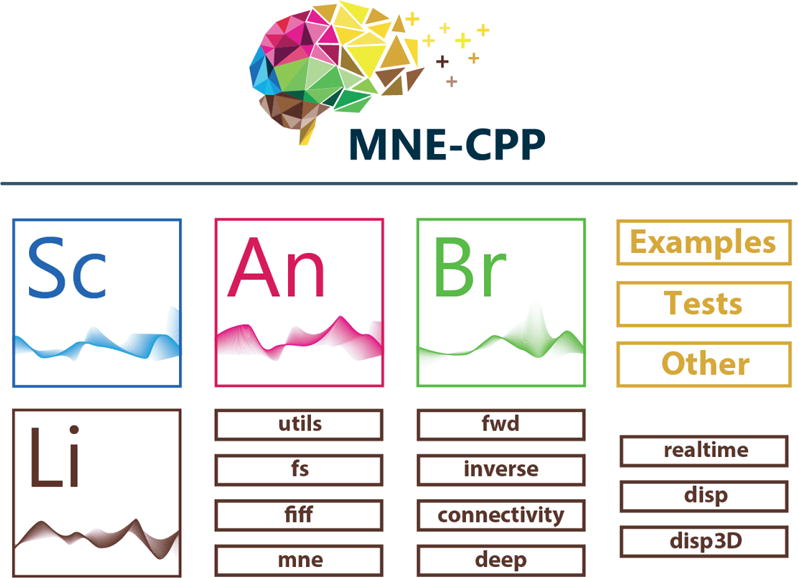
Overview of the MNE-CPP basic architectural layout, including application and library layer. Next to MNE Scan (Sc) there are several other applications which are built upon the MNE-CPP library layer (Li): MNE Browse (Br), MNE Analyze (An), to name a few.
MNE-CPP and MNE Scan make use of one external dependency, namely the Qt framework [23]. Also, we use a so-called Clone-and-Own approach of the Eigen library [24], which is integrated in our repository. Qt provides tools for GUI creation whereas Eigen provides mathematical operations for linear algebra. Minimizing the use of Software of Unknown Provenance (SOUP) is favorable when developing medical software applications meeting regulatory requirements: each third-party dependency must be tracked, and its development life cycle must be auditable. Furthermore, all dependencies should be able to compile on multiple platforms and devices for cross-platform capability.
2.2. Software Architecture Design Description
The architecture of MNE Scan is modular. The main application consists of five main classes. Most of the functionality is contained in MNE Scan’s shared libraries: scShared, scMeas, scDisp and its plugins. The scShared library holds all mechanisms to manage the plug-in creation, communication, and visualization. The real-time visualization and measurement data handling tools are implemented in the scDisp and scMeas libraries, respectively. This modularity ensures maintainable Application Programming Interfaces (APIs) and leads to a clear documentation structure for the medical device approval process.
The Unified Modelling Language (UML) class diagram in Figure 2 shows the key components of MNE Scan’s architecture. The three key manager classes are:
The PluginManager loads and instantiates plug-in object prototypes. Plug-in prototypes are created right after the shared plug-in objects are loaded at the startup of MNE Scan.
The PluginSceneManager holds plug-ins which were attached to PluginScene in the PluginGui via a PluginItem. Plug-ins are attached to the PluginGui by user interaction via PluginItems. The plugin interface provides a factory pattern, i.e., attached plug-ins are cloned of the prototypes and added to the PluginSceneManager via addPlugins(). The PluginSceneManager also starts and stops plug-ins with the startPlugins() and stopPlugins() methods respectively.
The DisplayManager attaches and returns the suitable MeasurementWidget to the given PluginOutputConnectors of the current plug-in which was selected with the respective PluginItem. The suitable MeasurementWidget is determined by the Measurement type associated with the PluginOutputConnector.
Figure 2.
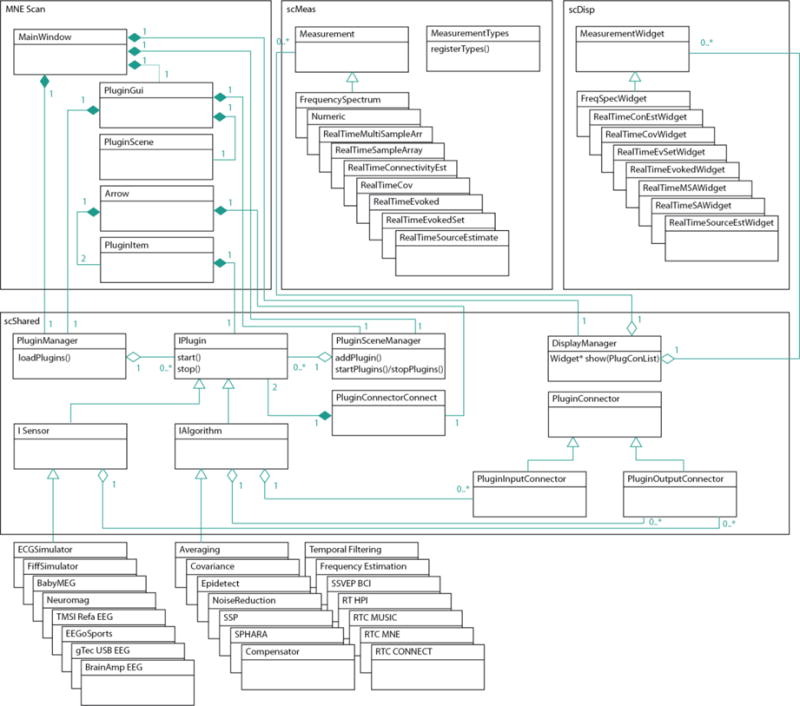
MNE-Scan UML class diagram with the most important architectural components. The main application MNE Scan consists of only a few classes which connect MNE Scan’s managers. MNE Scan Shared (scShared) library contains the definition of the managers as well as the plug-in interfaces of the sensors (ISensors) and algorithms (IAlgorithm). MNE Scan Measurement (scMeas) library contains the measurement types and MNE Scan Display (scDisp) library the corresponding measurement visualizations.
The data stream is realized with a PluginConnectorConnection object which establishes a connection with a user drawn Arrow between two plug-ins and their respective PluginOutputConnector and PluginInputConnector. The streaming data type is determined by the Measurement associated with both, input and output connectors. MNE Scan provides a variety of MeasurementTypes, see Table 2.
Table 2.
The different MeasurementTypes included in MNE Scan. Each type can hold and broadcast one specific data type.
| MeasurementType | Explanation |
|---|---|
| FrequencySpectrum | Spectrogram including header information |
| Numeric | Single floating-point value |
| RealTimeSampleArray | Single channel measurement of floating point values |
| RealTimeMultiSampleArray | Multi-channel measurement of floating point values |
| RealTimeConnectivityEstimate | Connectivity estimations |
| RealTimeCov | Covariance matrix estimations |
| RealTimeEvoked | Evoked data, i.e., an averaged or single trial section around a stimulus |
| RealTimeEvokedSet | An evoked data set, i.e., a set of averaged or single trial sections around multiple stimuli |
| RealTimeSourceEstimates | Source estimation data |
MNE Scan accepts two kinds of plug-ins: Algorithm and Sensor plug-ins. Sensor plug-ins can only have output connectors, whereas Algorithms can have both output and input connectors. Plug-ins are described in detail in Section 2.3.
The plug-in architecture implements the actual signal-processing functionality starting from device connectors through data acquisition to real-time processing. The sensor plug-ins can establish a data connection to external devices or already recorded data sets. The modular architecture allows creating processing pipelines, which are flexible and can differ between research and clinical plug-ins. The user interface for plug-in arrangement is described in Section 2.5.
2.3. The Plug-Ins
Corresponding to the chosen plug-in connections, the data are passed and processed throughout the user-defined pipeline. The raw data streams are created by the sensor plug-ins, i.e., by connecting to a measurement device such as EEG and MEG hardware. The received data are forwarded to the attached algorithm plug-ins where real-time processing tools are implemented. This way one creates an user-defined pipeline capable of providing a set of analysis data during the ongoing measurement session.
Each plug-in can be understood as a single functional unit, running in its own thread to perform multiple instructions in parallel. Data handling between the plug-ins is described in Section 2.5. The individual plug-ins are realized as separate libraries, which allows easy and efficient maintenance. To this point MNE Scan is equipped with seven sensor and twelve algorithm plug-ins.
Table 3 gives an overview of all currently available plug-ins and the corresponding input and output data types.
Table 3.
Available plug-ins in MNE Scan with corresponding input and output data streams as well as a description. DS = Data Stream, PP = Pre-Processed, RT = Real-Time, RTC = Real-Time Clustered.
| Plug-in Name | Input/Output | Description | |
|---|---|---|---|
| Sensor | BabyMEG | Device DS/Raw DS | Connects to the BabyMEG system. |
| Neuromag | Device DS/Raw DS | Connects to a Neuromag system. | |
| TMSI Refa EEG | Device DS/Raw DS | Connects to a TMSI amplifier. | |
| EEGoSports | Device DS/Raw DS | Connects to an EEGoSports amplifier. | |
| gTec USB EEG | Device DS/Raw DS | Connects to a gTec amplifier. | |
| BrainAmp EEG | Device DS/Raw DS | Connects to a BrainAmp amplifier. | |
| FiffSimulator | Pre-Rec. Data/Raw DS | Streams pre-recorded Fiff data. | |
| ECGSimulator | Pre-Rec. Data/Raw DS | Streams pre-recorded ECG data. | |
| Algorithm | SSP | Raw or PP DS/PP DS | Applies SSP.. |
| SPHARA | Raw or PP DS/PP DS | Applies SPHARA. | |
| Compensator | Raw or PP DS/PP DS | Applies software gradiometer. | |
| Temporal Filtering | Raw or PP DS/PP DS | Applies temporal/frequency filters. | |
| Averaging | Raw or PP DS/Averaged DS | Averages trigger based data. | |
| Covariance | Raw or PP DS/Covariance Estimation | Computes a covariance estimation. | |
| Frequency Est. | Raw or PP DS/Unchanged DS | Computes channel frequency spectra. | |
| SSVEP BCI | Raw or PP DS/Control Signal | SSVEP based BCI. | |
| RT HPI | Raw or PP DS/Head position determination | Estimates the head position in MEG. | |
| RTC MUSIC | Averaged DS/Source Estimation | Computes a MUSIC source estimate. | |
| RTC MNE | Averaged DS/Source Estimation | Computes a MNE source estimate. | |
| RTC CONNECT | Averaged DS/Connectivity Estimation | Computes a connectivity estimate. |
2.4. Data Handling
The connections and data handling between plug-ins and displays are realized by circular buffers implementing the observer design pattern [25]. Each time the maximum size of a buffer (subject) is reached the connected buffer clients (observers), e.g., plug-ins and displays, are notified. After being notified, the connected clients can copy the data from the buffer for further processing. Figure 3 depicts the data handling and communication in MNE Scan.
Figure 3.
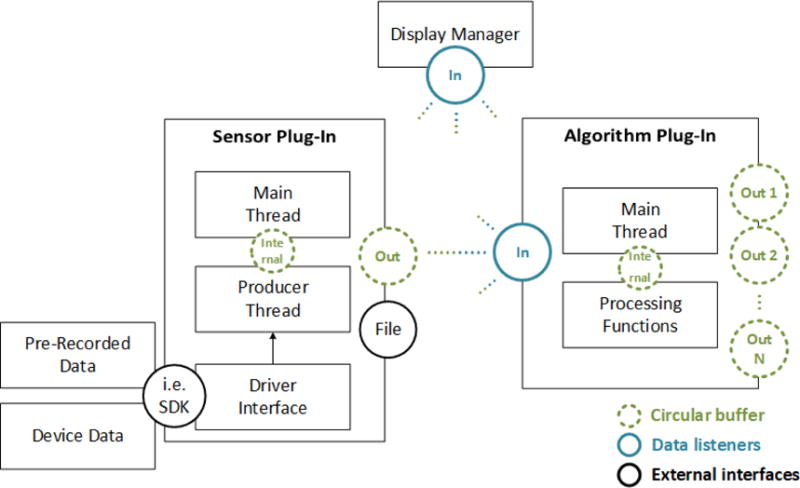
Data handling between plug-ins and visualization. The display manager handles the current data visualization depending on which plug-in was selected by the user.
Sensor plug-ins generate the raw data stream to be processed in real-time. If provided by the manufacturer, the raw data are collected using a device dependent Software Development Kit (SDK). For example, in case of the BabyMEG the related MNE Scan plug-in connects to an underlying LabView program [26]. By using a circular buffer, the received data blocks are forwarded from the driver and producer thread to the main thread. On this level the raw data blocks can be written to a file and streamed to algorithm plug-ins via an output buffer. Corresponding to the user-created pipeline and connections, multiple algorithm plug-ins can listen to the output buffer. Sensor plug-ins provide output data buffers. Algorithm plug-ins can listen to and provide an arbitrary number of buffers. This allows for fusion or extraction of information into a single or multiple output data streams.
To prevent data loss and unnecessary data read procedures, we introduce semaphores, which block data insertion into a buffer once it is full or read outs when no data are available. Only after data are pulled out of the buffer, the buffer will be freed for storing new data. This way no information is lost, which is a crucial requirement when dealing with patient data. In addition to the actual plug-ins, the display manager also listens to the currently selected plug-in and its attached output buffers. Consequently, the display manager provides the matching visualization for each output buffer in real-time. If a selected plug-in has multiple output buffers, the display manager creates an equal number of visualization widgets. MNE Scan is a multi-modal data processing application and therefore provides several different real-time structures to store data. The circular buffers described above are implemented as templates and can therefore handle different kind of data structures, see Table 2.
2.5. The Graphical User Interface
The GUI of MNE Scan is based on the widget concept in Qt, in which one main window holds multiple child widgets, which again can hold child widgets themselves. After startup, the user is presented with the main window of MNE Scan, which includes the plug-in manager, plug-in visualization and the main toolbar, see Figure 4. By default, the plug-in visualization shows a blank window if the pipeline has not been started yet. The plug-in scene is part of the plug-in manager and is used to position and connect the selected plug-ins. The connections are shown by arrows, which indicate the directions of data flow. The pipelines can be saved and loaded for later use. Sensor plug-ins are depicted as light blue and algorithm plug-ins as light green. The pipeline can be started or stopped using the Play or Stop button in the general toolbar. Figure 4 shows an example of an active pipeline, streaming a pre-recorded data set.
Figure 4.
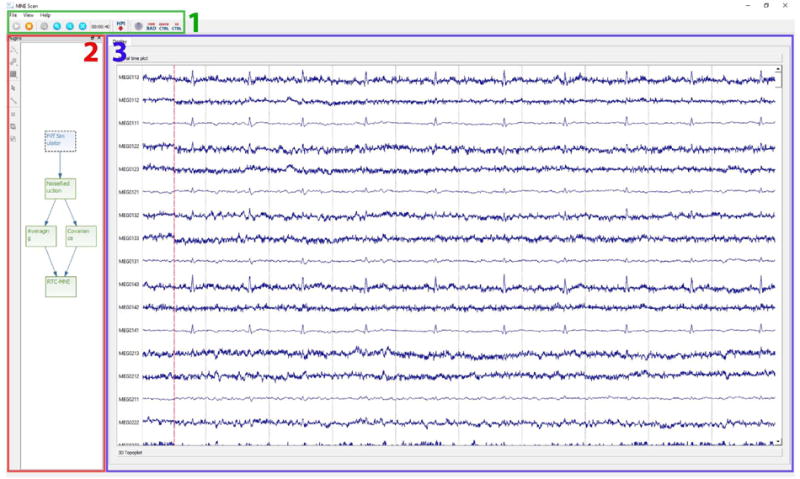
Screenshots of MNE Scan after the pipeline was started by the user: (1) The general toolbar is realized as a horizontal widget which holds the controls to start and stop the created pipeline as well as control widgets, e.g. the quick control widget or the channel selection manager. (2) The plug-in manager is framed in red and can be used to create the plug-in pipeline. (3) By clicking on the different plug-ins, the user is prompted with the corresponding visualization located in the display widget, framed in blue.
2.6. Real-Time Data Visualization
As described above, the display manager provides visualizations matching the currently selected plug-in and its output data buffers. Based on the data type in each outgoing buffer, the display manager creates a visualization widget and passes the buffer data to it. Real-time visualizations described below are part of the scDisp library and are presented in more detail in conjunction with use case examples in Section 4.
The RealTimeSAWidget, see Figure 2, for handling vector structures plots a single vector in one view, which provides freezing and tools for measuring amplitude and time scale of the plotted signal. The RealTimeMSAWidget for handling matrix structures plots multiple vectors in one view. The implementation is based on the Model-View-Controller (MVC) software architecture. Next to the row-wise signal course plotting it also features a 3D topographic plot. The RealTimeEvokedWidget and RealTimeEvokedSetWidget feature butterfly and 2D layout plots. The butterfly plot visualizes all selected channels from the selection manager and activated modalities on top of each other. The 2D layout plot makes use of a map of the channel locations and presents data time-locked to an event. A quick control widget is available for channel scaling, trigger marking and detection, screenshot taking, view and color adjustments. Channels of interest can be selected using the channel selection manager. The manager lets the user define groups of channels and store the selection to file for later or external use. EEG layouts are created on the fly via a 2D projection of 3D tracked electrode positions. The display also features noise reduction tools such as time-domain filtering, Signal Space Projection (SSP) [27], and synthetic gradiometer filtering. The noise reduction tools are only applied to the data stream send to the display widget, not the one sent to the subsequent connected plug-ins.
The RealTimeSourceEstWidget and RealTimeConEstWidget show estimated cortical activity and functional networks in real-time. The 3D models can be rotated and translated automatically or freely by the user. The real-time 3D visualization is based on the disp3D library of MNE-CPP, which internally relies on the Qt3D module. Using a 3D control widget, the user can choose from a variety of visualization options, such as real-time cortical activity smoothing, histogram thresholding, and colormap selection.
2.7. Performance
MNE Scan’s performance depends on several parameters including the sampling frequency, block size, number of measurement channels and the processing as well as visualization complexity of the chosen pipeline. To demonstrate these dependencies, we describe the performance of a pipeline including a basic data acquisition, noise reduction, averaging, covariance and source localization plug-ins. The pipeline and its results are described in more detail in section 4 and Figure 11. The performance timings were computed based on a 30 minute long test session. The test session implements the FiffSimulator plugin with pre-recorded data. The data were recorded with a Elekta Neuromag® MEG system with a total of 375 independent MEG, EEG and trigger channels at a sampling frequency of 600Hz. We examined three scenarios with the same data set streamed with different block sizes (100sp, 300sp and 600sp), resulting in different performance requirements for each step in the pipeline. In example, choosing a block size of 100sp at a 600Hz sampling frequency results in a maximum allowed processing time of 166ms. Not meeting this time requirement would introduce an overall delay and drag down the processing pipeline. All performance measurements were realized on a Dell XPS Laptop with an i7700HQ CPU, 16GB RAM, 512GB SSD and a NVIDIA GTX 1050M graphics card.
Figure 11.
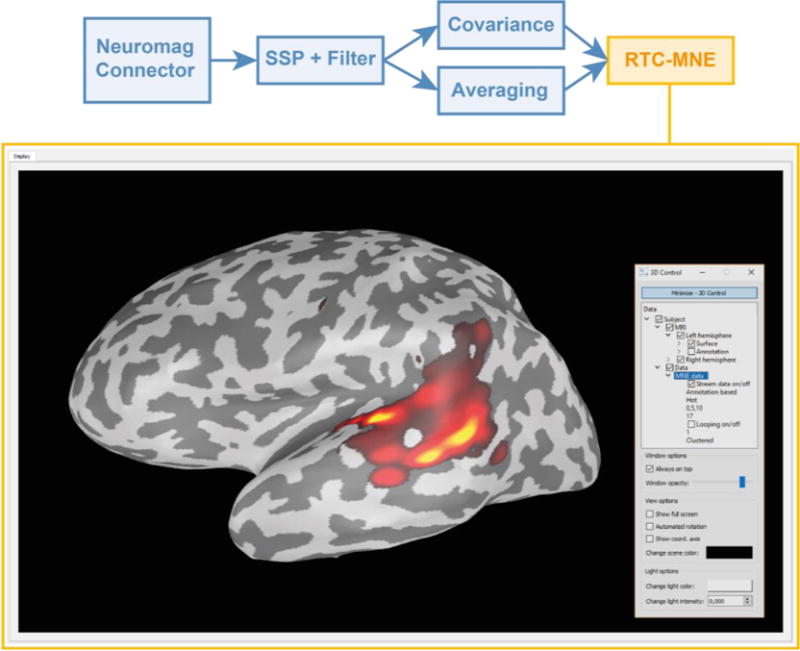
The 3D real-time visualization of a RTC-MNE estimate at 100 ms for 10 ten left auditory stimuli in form of a moving average. The control widget to the right can be used to change 3D visualization parameters during the ongoing measurement.
In Figure 5, the performance timings are shown for data handling in between plug-ins and internal data processing in each plug-in. Please note that the two tasks Averaging: Processing and Covariance: Processing are performed in independent threads and not subsequent to each other. Their results are then forwarded to the RTC-MNE plugin, see Figure 11.
Figure 5.
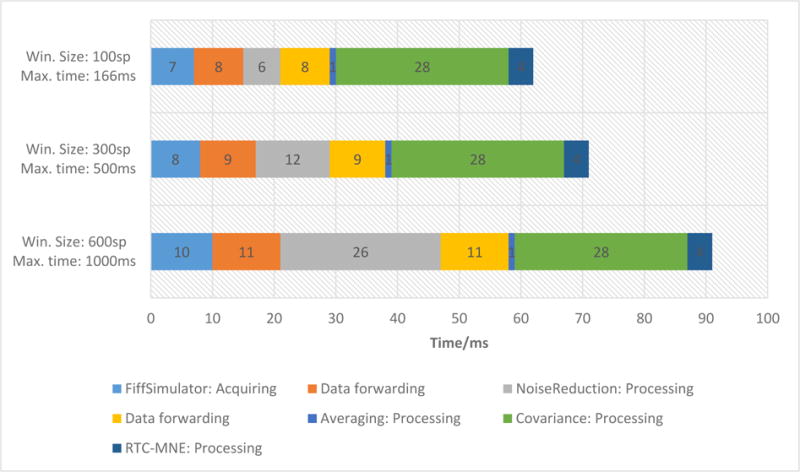
The performance timing for each processing and data handling task in the chosen pipeline. All performance timings are described in milliseconds. The three scenarios correspond to different block sizes (100sp, 300sp and 600sp) and maximum allowed time limits (166ms, 500ms, 1000ms). Each scenario was performed with a sampling frequency of 600Hz.
Figure 6 illustrates the performance timings for the result visualization of each plug-in. These timings are described separated from Figure 5 since the user can only visualize the results of one plug-in at a time. The timings in Figure 6 are based on the current visualization chosen by the user. Therefore, they are to be added to the data handling and processing timings in Figure 5 to obtain the complete timings for processing, data handling and chosen visualization. Please note that the Averaging and Covariance plug-ins work on buffered data, meaning they work on same data sizes in all three scenarios leading to equal performance values.
Figure 6.
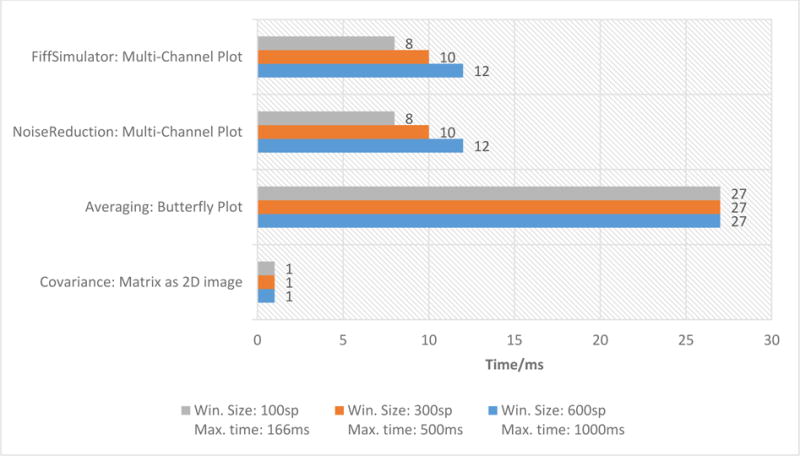
The performance timings for each plug-in’s result visualization tasks in the chosen pipeline. All performance values are described in milliseconds.
3. The Development Process
MNE Scan is developed within a strict software design control process, which is a requirement to be able to apply for clinical clearance [20]. The main motivation to undergo the approval process is to ensure that the risk of misinterpretation and therefore harming the patient is minimized. In consequence, the architecture should be as modular as possible to ensure that non-certified research modules do not affect essential clinical parts of the software. Therefore, MNE Scan pursues a strictly modular architecture, which also allows for fast prototyping of new methods that can first be tested in research studies. After the new methods are proven and medical approval has been obtained, the novel methods can be integrated into clinical workflow.
The MNE-CPP development process follows an iterative V-Model [20, 28] development approach. Figure 7 illustrates the main activities during the software development lifecycle. The left side of the V describes the path from establishment of the requirements to their implementation. The right side of the V includes their integration, verification, and validation. The V-Model development process was slightly changed to our specific needs, i.e., verification and validation activities of the V’s left side are already performed before the actual implementation, indicated by horizontal lines in Figure 7. Design concepts can, e.g., be verified with mockup studies. The decomposition in separate activity steps provides guidance for the planning and realization of standardized development processes. This ensures the traceability of software quality. Furthermore, the V-Model improves the communication between all project stakeholders, i.e., the clinical and the developers side, and minimizes project risk.
Figure 7.
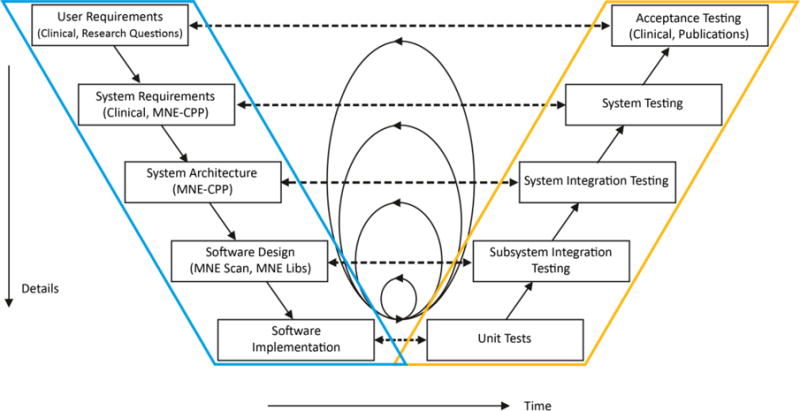
MNE-CPP’s system development process follows the V-Model. Within the model the key activities are shown. The circles in the middle represent the iterative approach, namely the agile development iterations with SCRUM. The area framed in blue includes all implementation efforts. The yellow area represents software testing efforts.
The MNE-CPP project iterates in loops within the V-Model using the SCRUM project management methodology to be able to quickly react to new and unforeseen requirements. SCRUM is an agile project management technique, where short time periods are organized as one internal project cycle, after which a fixed number of tasks should be undertaken to produce a working prototype. We use JIRA, a well-established task tracking and project management platform, see https://mne-cpp.atlassian.net.
4. Use-Case Scenarios
To present and evaluate MNE Scan’s real-time capabilities, we describe three use-case scenarios. The first is closely connected to a clinical study with the BabyMEG system. In the second use case, we describe the results from the integration of a more research-oriented setup with novel real-time source estimation methods being investigated. Finally, we present a proof of principle implementation of an EEG BCI speller.
Clinical Epilepsy Study
The BabyMEG [29] is a 375-channel whole-head pediatric MEG system. It is setup in a clinical environment at Boston Children’s Hospital to study inpatients and outpatients as well as healthy neonates, infants, and preschool children up to three years of age. MNE Scan is used to acquire data and to display the incoming data during the ongoing measurement session. The RealTimeMSAWidget was used to visualize the incoming data. For the BabyMEG to operate outside of basic scientific areas and be used in daily clinical use as well as patient studies, FDA approval was obtained for the hardware and software components. MNE Scan’s BabyMEG plug-in connects to these approved components. Figure 8 shows a snapshot of the BabyMEG plug-in in action during a measurement.
Figure 8.
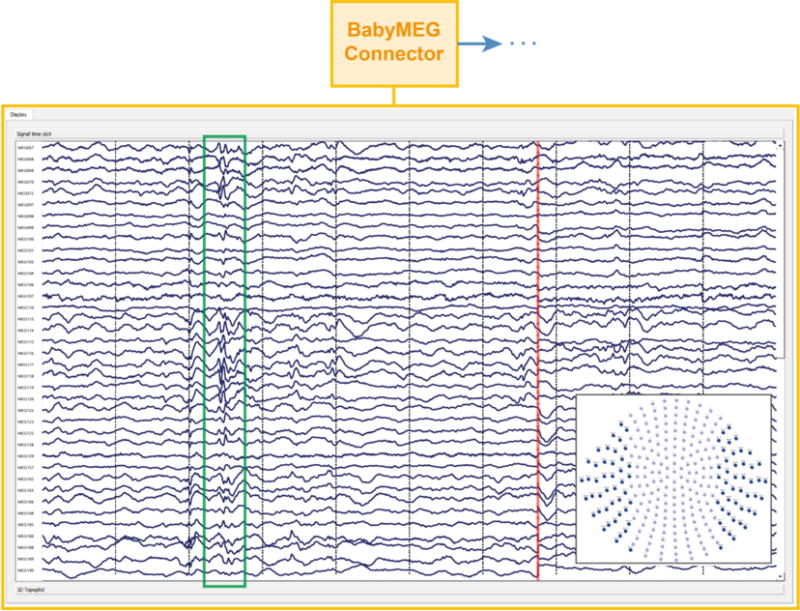
The FDA approved main MNE Scan application and BabyMEG plug-in. The snapshot shows the real-time visualization of the incoming data stream of left and right temporal MEG channels during a clinical epilepsy study. The epileptic activity is marked by the green rectangle. The end of the last received data block is indicated by the red vertical line. Further connection of algorithm plug-ins is implied by the dots next to the arrow in the upper part of the figure.
Next to viewing the data in real-time, the BabyMEG plug-in offers file writing, a basic subject manager and timing features. The external magnetic field interference was removed in real time and the spontaneous brain activity including the epileptiform activity was displayed on a monitor online for immediate analysis of the epileptiform activity of the patient. The green rectangle in Figure 8 indicates spontaneous epileptic activity measured from a nine-month-old patient. Please note, that the spike marking was not performed in real-time, but instead was added to the figure as an overlay afterwards. Subsequent algorithm plug-ins can be connected to the BabyMEG plug-in.
Real-time source estimation
The chosen pipeline made use of all major algorithm plug-ins included in MNE Scan. The data were recorded with the Neuromag sensor plug-in. The subject was presented with left auditory stimuli. Again, the data blocks were visualized with the RealTimeMSAWidget. Subsequently, the MEG data were pre-processed using the Temporal Filtering and SSP plug-ins, see Figure 9.
Figure 9.
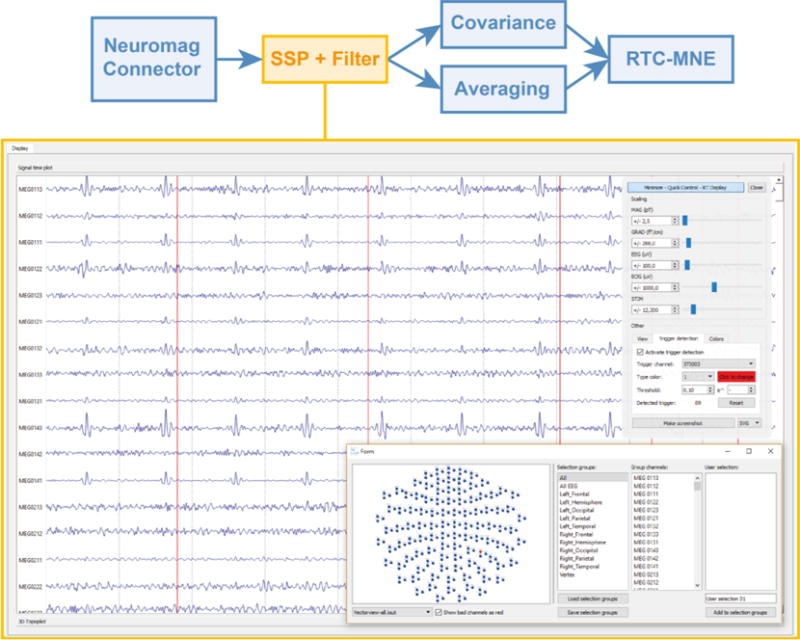
Real-time visualization of the data stream after it was received from the Neuromag plug-in and processed in the SSP and temporal filter plug-ins. The vertical red lines indicate the detected triggers for each auditory stimulus. The quick control widget and selection manager are shown in the upper and lower right part of the figure, respectively. Subsequently, the data stream is forwarded to the covariance and averaging plug-ins, which run in parallel to each other.
The averaging plug-in detected triggers occurring in a selected trigger channel, cut out the correct time segment and generated moving averages. Each average thereby consisted out of ten trials ranging from -100 ms to 400 ms, relative to trigger onset. The computed averages were visualized as butterfly plots, see Figure 10. The resulting pre-processed data stream was forwarded to the averaging and covariance plug-in. The covariance plug-in estimated the noise covariance matrix [4], which was needed in subsequent algorithm plug-ins.
Figure 10.
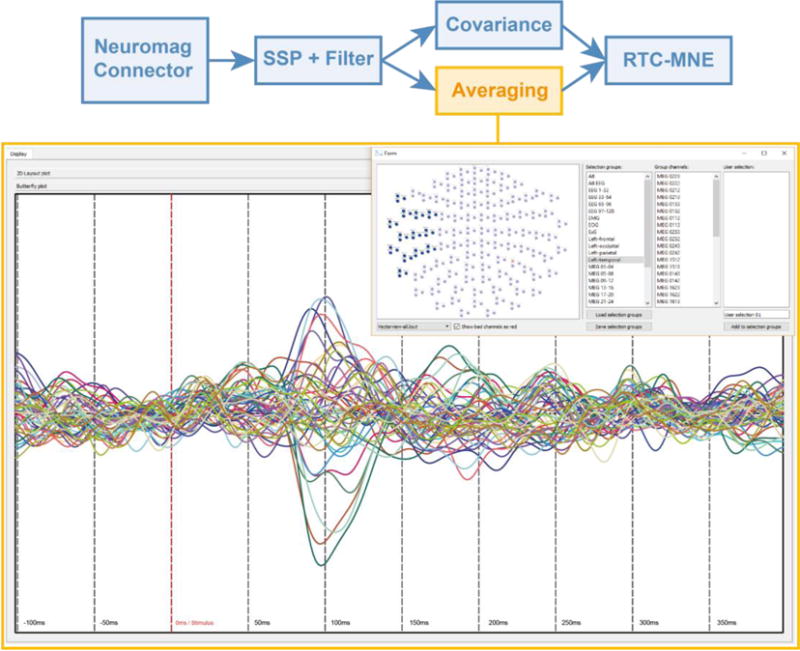
Real-time visualization of the averaging results in form of a butterfly plot. Using the selection manager only the channels of interest in the left occipital area were selected.
The averaged and noise covariance data were sent to the Real-Time Clustered - Minimum Norm Estimates (RTC-MNE) [30] plug-in, which provides a distributed source estimate of neuronal activity. The covariance estimate was used to take the different noise levels and correlations into account in the construction of the inverse operator. A sparse source estimate can be calculated by the Real-Time Clustered - Multiple Signal Classification (RTC-MUSIC) [31] plug-in as well. RTC-MUSIC generates a sparse representation of the most correlating dipoles. Both RTC-MNE and RTC-MUSIC are working on a clustered source space to reduce the computational burden and deal with a low signal to noise ratio (SNR). The estimated source activity was visualized on a freely rotatable 3D cortical surface reconstructed with the Freesurfer software [32]. Figure 11 illustrates the real-time visualization of a RTC-MNE result on top of an inflated cortical surface.
EEG BCI speller
As part of a proof of principle study we implemented a steady-state-visual-evoked-potential (SSVEP) based BCI in MNE Scan. The implementation was partially adapted from the Bremen BCI [33]. SSVEP based BCIs require an external visual stimulation. Consequently, a visual reactive BCI was integrated into MNE Scan. As an example use case scenario, we chose a speller application where the user can steer through and select letters from a virtual keyboard. During test sessions subjects were equipped with dry EEG caps [34] connected to an EEGoSports amplifier, see Figure 12. right. The implementation included the necessary stimulation via five flashing rectangles, see Figure 12 left, all BCI processing steps in form of a single plug-in and the virtual keyboard application. Preprocessing steps were realized with the Temporal Filtering and SPHARA [35] plug-ins. The data acquisition was provided by the EEGoSports plug-in. The implemented BCI delivered an average detection accuracy of 86 %.
Figure 12.
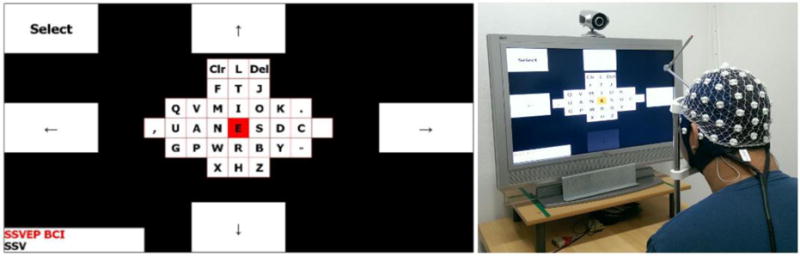
SSVEP BCI speller implemented in MNE Scan. Left: The flashing rectangles to steer through and select the virtual keyboard letters. Right: An example BCI session with a subject equipped with a dry EEG cap.
5. Discussion
The three major goals of MNE Scan were to provide software for real-time data analysis, to follow regulatory requirements, and to reduce the time between the development and introduction of the new tools to clinical studies.
Based on several use cases we were able to confirm the feasibility of MNE Scan’s real-time analysis tools. Real-time processing demands an efficient implementation. Even when using an efficient language such as C++, all program architectures, algorithms, and data structures must be optimized. Especially the fast visualization of large data sets is a challenge. All 2D visualizations in MNE Scan are currently realized on CPU level on single thread basis. Each Qt widget runs in its own thread. This can become a problem when the data are too large and there are many parallel processing tasks. Especially plug-ins with advanced visualizations, e.g., 3D topographic and source activity plots, are computationally costly. Visualizing and processing at high sampling rates (> 10 kHz) often used in invasive electrophysiological recordings are currently not supported when the number of channels is large (> 100). Importantly, standard sampling rates (< 2 kHz) employed in high-density EEG or whole-head MEG systems with hundreds of channels are well within the capabilities of MNE Scan, e.g., in combination with the BabyMEG’s hardware controller [26]. We have already identified some of the computational bottlenecks to be able to improve the performance. The plug-in result visualization can be further improved by, e.g., facilitating OpenGL programming. When working with modular units, such as plug-ins in MNE Scan, the efficiency of data communication between the units is of outmost importance. Improved circular buffers and semaphores are potential solutions. Providing current performance timings during the measurement session can give the user an imminent idea whether the chosen pipeline is in fact delivering real-time results or requires further adaption. The authors in [36] provide a tool for monitoring the instantaneous real-time capability of their software. We plan to add a similar tool to MNE Scan in the future.
MNE Scan internally relies on the Fiff (functional image file format) specification. Subsequently, other file formats and data structures must be mapped to the Fiff specification. By introducing plug-ins as individual units, research groups can tailor acquisition and processing to their individual needs. This enables acquisition systems and advanced real-time processing tools to be used in new research and clinical studies. With its open-source license MNE Scan can be freely accessed and adapted by the users. Significant effort has been invested in designing the code interfaces to be as user friendly as possible. Still, individualizing MNE Scan requires understanding of C++ and object-oriented programming.
MNE Scan demonstrates that it is feasible and even favorable to follow a strict development cycle in an open-source project. With MNE Scan we provide a modular architecture, keeping dependencies to a minimum which is beneficial for medical software approval. To our knowledge there are no other similar projects which consider the organizational, code, and testing requirements to obtain clinical software approval. The modular architecture ensures a minimal interference between plug-ins and shared libraries. Therefore, it can be anticipated that the medical approval of MNE Scan’s main application, its plug-ins and shared libraries can be performed modular as well. MNE Scan and MNE-CPP are still undergoing major development efforts. Contributions by the community are therefore crucial and encouraged.
MNE Scan and its real-time features were successfully tested in several use case scenarios. These use case scenarios included clinical studies (BabyMEG, epilepsy) and basic research (real-time source estimation and BCI). This demonstrates the ability of MNE Scan to allow state of the art methods to exist in the same ecosystem as clinically approved tools. MNE Scan is deployed in two ways. A user can build the binaries from the source code provided. In addition, we provide prebuilt binaries.
6. Summary and Outlook
As an acquisition and real-time processing software MNE Scan can control the electrophysiological hardware to acquire data. MNE Scan provides fast and effective algorithms to process the acquired data in real-time. It has a plug-in-based architecture to adapt and extend its MEG and EEG real-time processing capabilities. MNE Scan provides a platform for development, which allows new devices to be easily integrated into the device repertoire. This reduces the time between introduction of new devices and methods and their clinical use. Moreover, the development of MNE-CPP and MNE Scan follows strict regulatory requirements, which enables clinical clearance.
The development of MNE Scan will continue in the direction of a multi-purpose acquisition, processing and visualization platform, merging different measurement modalities that in future will include invasive electrophysiological measurements. We thus envision MNE Scan as the common platform for acquiring and integrating data from different electrophysiological measurements. For example, in clinical evaluation of epilepsy patients the data are today obtained and analyzed using a variety of software packages, requiring a significant investment of time in training. Data integration is difficult and often qualitative. Our unified approach using MNE Scan will not only reduce the cost of training technicians to collect and analyze various types of data, but we also anticipate changes in clinical practice by introducing integration of all modalities and by enabling new approaches in surgical management.
Highlights.
MNE Scan is a new software for acquiring and processing electrophysiological data in real-time.
This work is a first step in establishing a standardized real-time processing software targeting large parts of the neuroscience community.
The employed software development cycle considers the requirements needed for clinical software approval processes.
MNE Scan was tested in multiple real-time scenarios. It is in active use with a new pediatric Magnetoencephalography (MEG) system which was already approved by the Food and Drug Administration (U.S. FDA).
MNE Scan is developed under an open-source license and is freely available as source code or pre-built binaries.
Acknowledgments
At this point we would like to thank all developers for their contributions to the MNE Scan and MNE-CPP project. Also, we would like to thank Banu Ahtam, Ashley Curran and Katie Gill for providing BabyMEG data and software testing.
Funding
This work is supported by the National Institutes of Health [NIH, grants 5R01EB009048, 2P41EB015896, and 1U01EB023820]; Deutsche Forschungsgemeinschaft [DFG, grant DFG Ba 4858/1-1]; Boston Children’s Hospital; Technische Universität Ilmenau; and the German Academic Exchange Service (DAAD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: None
References
- 1.Cohen D. Magnetoencephalography: Evidence of Magnetic Fields Produced by Alpha-Rhythm Currents. Science (80-) 1968 Aug;161(3843):784–786. doi: 10.1126/science.161.3843.784. [DOI] [PubMed] [Google Scholar]
- 2.Hämäläinen MS, Ilmoniemi RJ. Interpreting magnetic fields of the brain: minimum norm estimates. Med Biol Eng Comput. 1994 Jan;32(1):35–42. doi: 10.1007/BF02512476. [DOI] [PubMed] [Google Scholar]
- 3.Berger H. Über das Elektrenkephalogramm des Menschen I to XIV. Arch Psychiatr. 1929 [Google Scholar]
- 4.Gramfort A, et al. MNE software for processing MEG and EEG data. Neuroimage. 2014;86:446–60. doi: 10.1016/j.neuroimage.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gramfort A, et al. MEG and EEG data analysis with MNE-Python. Front Neurosci. 2013 Dec 1–13;7:7. doi: 10.3389/fnins.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tadel F, Baillet S, Mosher JC, Pantazis D, Leahy MR. Brainstorm: A User-Friendly Application for MEG/EEG Analysis. Comput Intell Neurosci. 2011;2011:1–13. doi: 10.1155/2011/879716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delorme A, Makeig S. EEGLAB: an open source toolbox for analysis of single-trail EEG dynamics including independent component analysis. J Neurosci Methods. 2004;134:9–21. doi: 10.1016/j.jneumeth.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Oostenveld R, Fries P, Maris E, Schoffelen J-M. FieldTrip: Open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput Intell Neurosci. 2011;2011:156869. doi: 10.1155/2011/156869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gramfort A, Papadopoulo T, Olivi E, Clerc M. OpenMEEG: opensource software for quasistatic bioelectromagnetics. Biomed Eng Online. 2010;9:45. doi: 10.1186/1475-925X-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.BESA GmbH. BESA: Brain Electrical Source Analysis. 2017 [Online] Available: www.besa.de.
- 11.Compumedics NeuroScan. Curry. 2017 [Online] Available: http://compumedicsneuroscan.com.
- 12.ANT Neuro. asa. 2017 [Online] Available: https://www.ant-neuro.com/
- 13.EGI. EMSE. 2017 [Online] Available: www.egi.com.
- 14.Bergmann TO, Karabanov A, Hartwigsen G, Thielscher A, Roman RH. Combining non-invasive transcranial brain stimulation with neuroimaging and electrophysiology: Current approaches and future perspectives. Neuroimage. 2016;135 doi: 10.1016/j.neuroimage.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Bogner W, et al. 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage. 2014 Dec;103(7453):290–302. doi: 10.1016/j.neuroimage.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kothe CA, Makeig S. BCILAB: A platform for brain-computer interface development. J Neural Eng. 2013;10(5):56014. doi: 10.1088/1741-2560/10/5/056014. [DOI] [PubMed] [Google Scholar]
- 17.Schalk G, McFarland DJ, Hinterberger T, Birbaumer N, Wolpaw RJ. BCI2000: a general-purpose brain-computer interface (BCI) system. IEEE Trans Biomed Eng. 2004 Jun;51(6):1034–43. doi: 10.1109/TBME.2004.827072. [DOI] [PubMed] [Google Scholar]
- 18.Renard Y, et al. OpenViBE: An Open-Source Software Platform to Design, Test, and Use Brain–Computer Interfaces in Real and Virtual Environments. Presence. 2010;19(1):35–53. [Google Scholar]
- 19.OpenBCI. OpenBCI. 2017 [Online] Available: www.openbci.com.
- 20.IEC. IEC 62304:2006+AMD1:2015 Medical device software - Software life cycle processes. 2015:170. [Google Scholar]
- 21.Dinh C, Luessi M, Sun L, Haueisen J, Hamalainen SM. MNE-X: MEG/EEG Real-Time Acquisition, Real-Time Processing, and Real-Time Source Localization Framework. Biomed Eng (NY) 2013 Sep;58(1):4184. doi: 10.1515/bmt-2013-4184. [DOI] [PubMed] [Google Scholar]
- 22.Dinh C. Brain Monitoring: Real-Time Localization of Neuronal Activity. 1st. Aachen; Shaker: 2015. [Google Scholar]
- 23.The Qt Company, Digia Plc, Nokia, and Trolltech. Qt 5.7.1. Oslo: 2016. [Google Scholar]
- 24.Guennebaud G, Jacob B. Eigen v3. 2010 [Online] Available: http://eigen.tuxfamily.org.
- 25.Gamma E, Helm R, Johnson R, Vlissides J. Design Patterns – Elements of Reusable Object-Oriented Software. 2002 [Google Scholar]
- 26.Sun L, et al. Versatile synchronized real-time controller for large-scale fast data acquisition. Rev Sci Instrum. 2017;88(5) doi: 10.1063/1.4983080. [DOI] [PubMed] [Google Scholar]
- 27.Uusitalo MA, Ilmoniemi RJ. Signal-space projection method for separating MEG or EEG into components. Med Biol Eng Comput. 1997;35(2):135–140. doi: 10.1007/BF02534144. [DOI] [PubMed] [Google Scholar]
- 28.German Directive 250, Software Development Standard for the German Federal Armed Forces, V-Model, Software Lifecycle Process Model, August 1992. 1992 [Google Scholar]
- 29.Okada Y, et al. BabyMEG: A whole-head pediatric magnetoencephalography system for human brain development research. Rev Sci Instrum. 2016 Sep;87(9):94301. doi: 10.1063/1.4962020. [DOI] [PubMed] [Google Scholar]
- 30.Dinh C, et al. Real-Time MEG Source Localization Using Regional Clustering. Brain Topogr. 2015;28(6):771–784. doi: 10.1007/s10548-015-0431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dinh C, et al. Real-Time Clustered Multiple Signal Classification (RTC-MUSIC) Brain Topogr. 2017 doi: 10.1007/s10548-017-0586-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischl B. FreeSurfer. Neuroimage. 2012 Aug;62(2):774–81. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valbuena D, Sugiarto I, Gräser A. Spelling with the Bremen brain-computer interface and the integrated SSVEP stimulator. Proc 4th Int Brain-Computer Interface Work Train Course. 2008 Sep;:291–296. [Google Scholar]
- 34.Fiedler P, et al. Novel Multipin Electrode Cap System for Dry Electroencephalography. Brain Topogr. 2015;28(5):647–656. doi: 10.1007/s10548-015-0435-5. [DOI] [PubMed] [Google Scholar]
- 35.Graichen U, Eichardt R, Fiedler P, Strohmeier D, Zanow F, Haueisen J. SPHARA - A Generalized Spatial Fourier Analysis for Multi-Sensor Systems with Non-Uniformly Arranged Sensors: Application to EEG. PLoS One. 2015;10:1–22. doi: 10.1371/journal.pone.0121741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel YA, George A, Dorval AD, White JA, Christini DJ, Butera JR. Hard real-time closed-loop electrophysiology with the Real-Time eXperiment Interface (RTXI) PLoS Comput Biol. 2017;13(5):1–22. doi: 10.1371/journal.pcbi.1005430. [DOI] [PMC free article] [PubMed] [Google Scholar]


