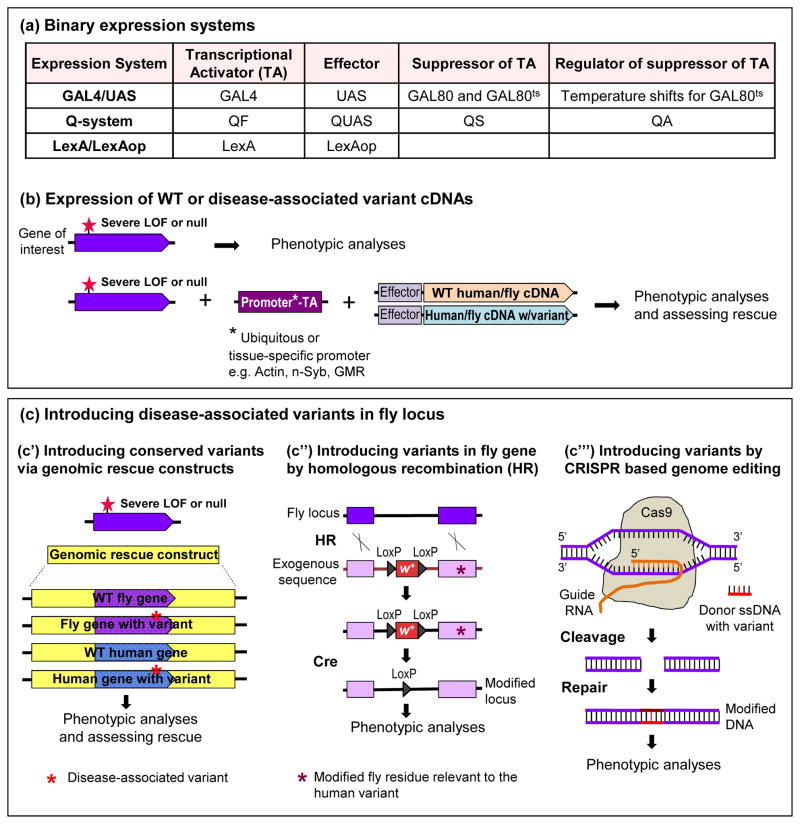
FIGURE 2.
Methods to model Drosophila for assessing disease variants (a) Basic binary expression systems used in Drosophila (b) Upon identification of phenotypes caused by a severe LOF mutation, the effect of overexpression of WT or disease-variant bearing cDNA (of both fly and human) can be tested using binary expression systems: GAL4/UAS, Q-system, and LexA/LexAop. Availability of a wide-range of tissue/developmental stage-specific drivers and conditional use of binary systems allow elegant manipulations on gene expression. (c) Disease-associated variants can be inserted into genomic rescue constructs or into the endogenous fly locus when the variant is conserved in the fly homologue. (c′) If the gene of interest contains a single exon, the fly exon can be replaced by wild type or disease variant-containing human exon. Introducing these genomic rescue constructs into flies that lack the gene of interest and display certain phenotypes allows testing for rescue with different genomic constructs. (c″) Exogenous fly sequence with the disease variant can be replaced by the endogenous fly locus using ends-out homologous recombination (HR). Cre recombinase induces site-specific recombination between LoxP sites that flank mini white marker (w+) and removes w+, leaving a LoxP within the intron. This method can be used to create knock in disease models in flies. (c‴) CRISPR/Cas9 based genome editing can be used to precisely modify nucleotides. RNA-guided Cas9 nuclease induces a double-strand break. Homology directed repair repairs the break by introduction of a donor single-stranded DNA (ssDNA) containing disease-variant.