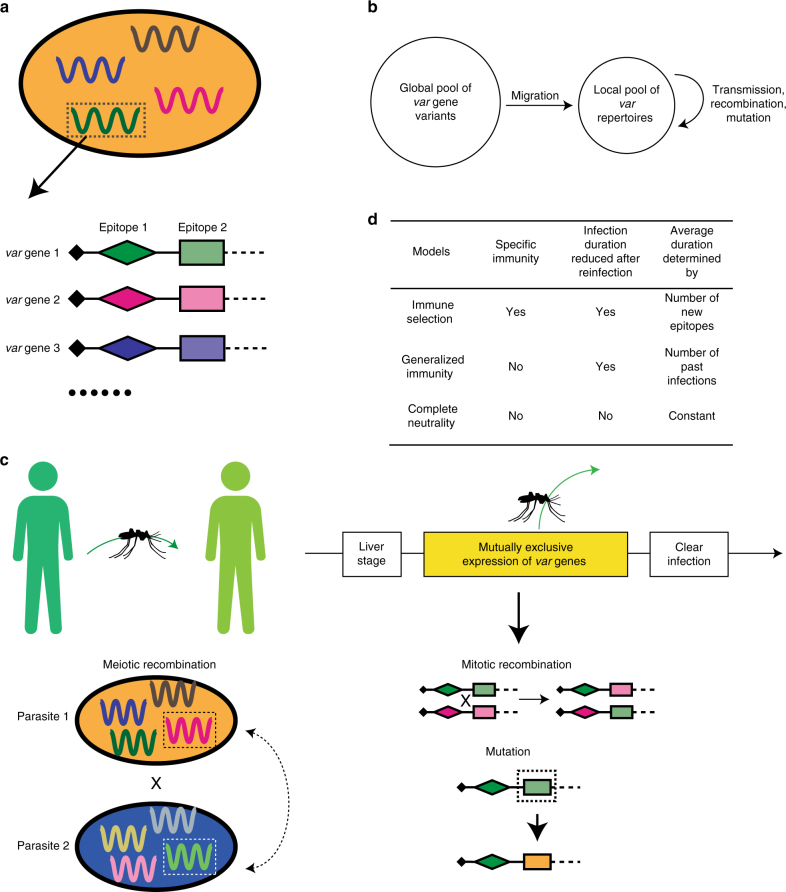
Fig. 1.
Schematic illustration of the var gene model. a Each parasite genome (ovals) consists of a repertoire of g copies of var genes. Each var gene (depicted by different colors within each parasite) is in turn represented as a linear combination of epitopes (depicted by different shapes), with each epitope having many possible variants (alleles, depicted by different colors). b The local population receives var repertoires from a fixed global var gene pool through migration. c At each transmission event, one donor and one receiver host are selected at random from the host pool. Each parasite genome in the donor host is transmitted to the mosquito with probability of 1/(number of genomes). During the sexual stage of the parasite (within mosquitoes), different parasite genomes can exchange var repertoires through meiotic recombination to generate novel recombinant repertoires. The receiver host can receive either recombinant genomes or original genomes. During the asexual reproduction stage of the parasite (within the blood stage of infection), var genes within the same genome exchange epitope alleles through mitotic (ectopic) recombination. Also, epitopes can mutate. These two processes generate new var genes. Each var gene is expressed sequentially and the infection ends when all the var genes in the repertoires have been expressed. A new transmission event may occur throughout the period of expression of var genes as the result of biting events. d Comparison of the three different models. Only the immune selection model includes specific immunity, through the dependence of infection duration on the memory of previous alleles that have been seen by a given host. For meaningful comparisons, all the models have the same mean duration of infection