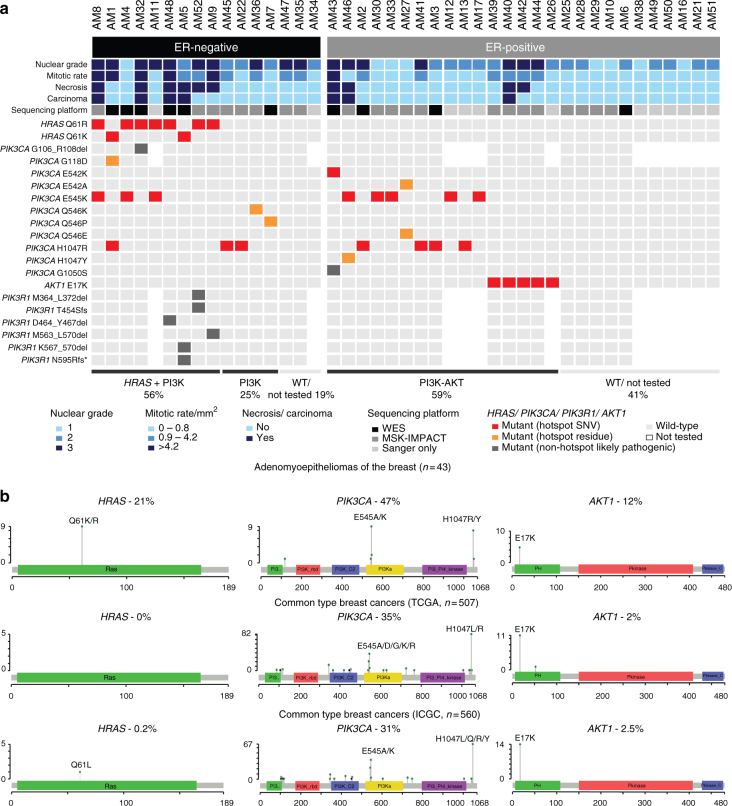
Fig. 3.
Somatic mutations affecting HRAS and PI3K-AKT pathway-related genes in breast adenomyoepitheliomas. a Heatmap depicting somatic mutations affecting HRAS, PIK3CA, AKT1, and PIK3R1 identified in 43 breast adenomyoepitheliomas by (i) both massively parallel sequencing (WES or MSK-IMPACT) and Sanger sequencing or (ii) Sanger sequencing only. Cases are shown in columns (estrogen receptor (ER)-negative cases on the left; ER-positive cases on the right), and genes in rows. Histopathologic characteristics and sequencing platforms are shown at the top, and color-coded according to the legend at the bottom. Hotspot mutations were obtained from Chang et al.57. SNV, single nucleotide variant; WES, whole-exome sequencing; WT, wild-type. b Spectrum of somatic mutations affecting HRAS, PIK3CA, and AKT1 identified in the 43 breast adenomyoepitheliomas analyzed in this study, and in unselected breast cancers from The Cancer Genome Atlas (TCGA, n = 507)6 and International Cancer Genome Consortium (ICGC, n = 560)7 studies. Diagrams representing the protein domains of HRAS encoded by HRAS (left), p110α encoded by PIK3CA (middle), and AKT1 encoded by AKT1 (right). The mutations in these three genes are shown on the x-axis, and the height of each “lollipop” indicates the frequency of the mutation (y-axis). Missense mutations are depicted as green circles, and small insertions and deletions are shown in black circles. Plots were generated using MutationMapper on cBioPortal (www.cBioPortal.org) and were manually edited