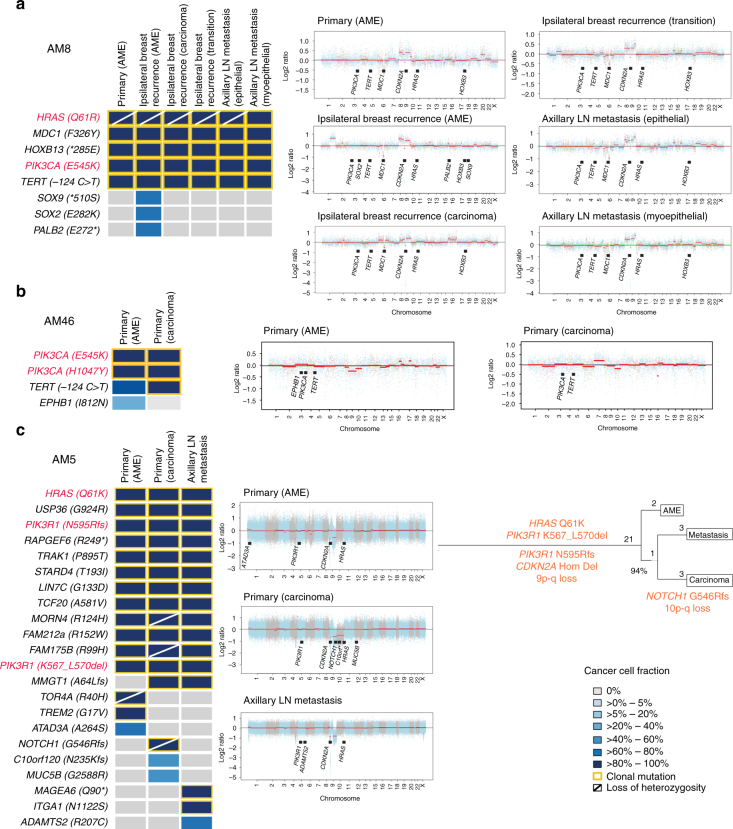
Fig. 4.
Progression of breast adenomyoepitheliomas. a−c On the left, heatmaps depicting the cancer cell fractions of the mutations identified in separately microdissected, histologically distinct components of AM8, AM46, and AM5; and on the right the copy number genome plots of each component. Cancer cell fractions were determined using ABSOLUTE12, and color-coded according to the legend. Loss of heterozygosity (LOH) is shown with a diagonal bar. Clonal mutations are highlighted with orange boxes. Mutations affecting HRAS and/or PI3K-AKT pathway-related genes are highlighted in red. In copy number genome plots, the genomic position is plotted on the x-axis and the Log2 ratios on the y-axis, and the cancer genes affected by somatic mutations and any gene affected by subclonal somatic mutations are shown according to their genomic position. a AM8, where the primary adenomyoepithelioma (AME), separately microdissected components of an ipsilateral relapse in the breast tissue (AME, invasive ductal carcinoma and transition components) and of separately microdissected components of a metachronous axillary lymph node metastasis (epithelial- and myoepithelial-enriched components) were analyzed by MSK-IMPACT sequencing. b AM46, where the breast adenomyoepithelioma and spindle cell metaplastic carcinoma components of the primary tumor were analyzed by MSK-IMPACT sequencing. c AM5, where the breast adenomyoepithelioma and myoepithelial metaplastic carcinoma components of the primary tumor and a synchronous axillary lymph node metastasis were analyzed by whole-exome sequencing (WES). The phylogenetic tree was constructed using Treeomics62. The length of the trunk and branches is proportional to the number of mutations defining each trunk or branches. Likely driver genes and copy number alterations found in the trunk and branches are highlighted in orange. Hom Del, homozygous deletion; LN, lymph node