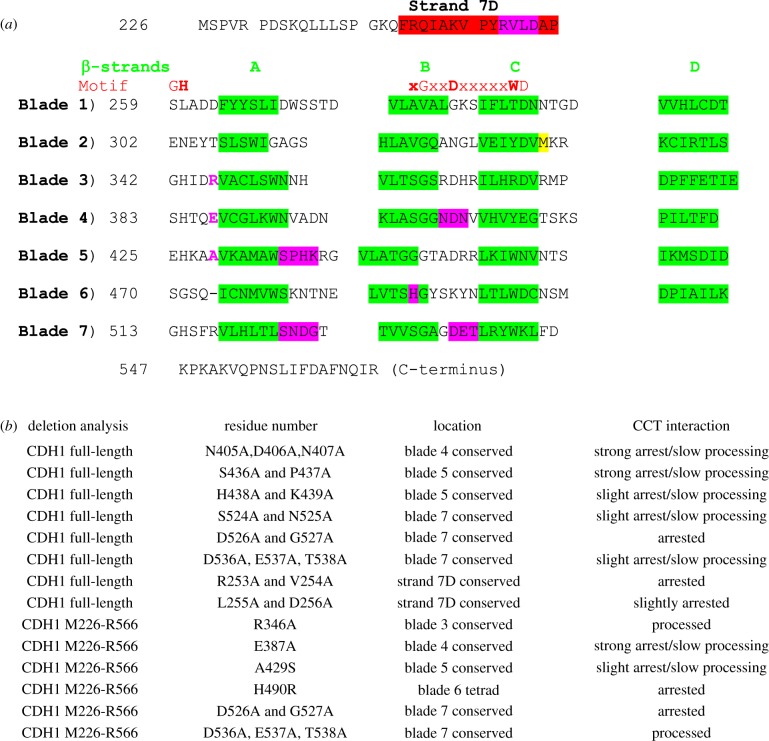
Figure 4.
Mapping CCT-binding residues in the Cdh1p propeller. Mutants in the Cdh1p propeller (amino acid residues 226–566) were screened for strength of binding and degree of processing by CCT using in vitro translation/native gel analysis. Six mutations were tested in the context of the propeller domain only. (a) The β-strands in the blades are highlighted in green. Motif: the red bold letters highlight the residues (GH-G-D-WD) forming the hydrogen-bonding network (‘structural tetrad’) which stabilizes the blades. The M332A mutation, coloured yellow, is included to prevent internal translation initiation at this methionine which reduces the accuracy of quantitation in these assays [27]; the equivalent residue in human CDH1 is alanine. The point mutants which arrest the processing are highlighted in magenta. (b) List of 14 mutants and their CCT interaction behaviour. Strong arrest indicates that greater than 15% of the counts become associated with CCT; slow processing means that the appearance of the folded Cdh1 (either full-length or the propeller domain) is retarded compared to wild-type control.