Fig. 7.
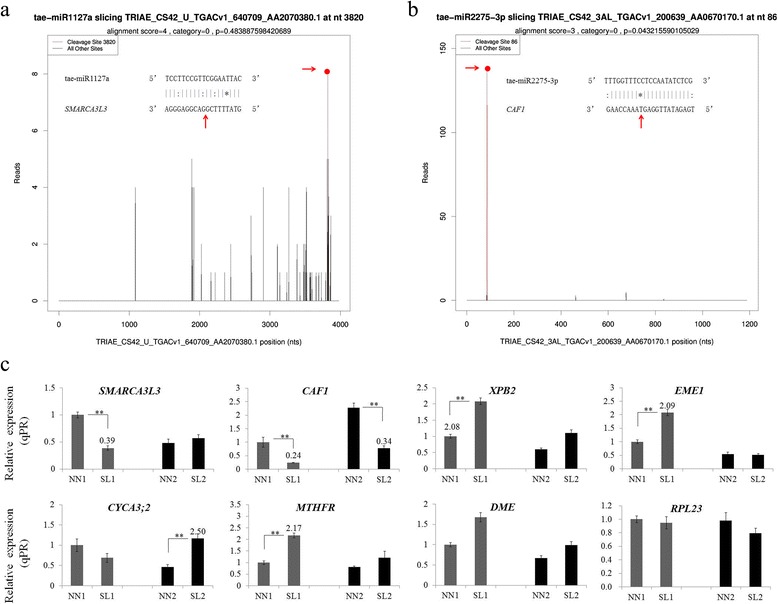
T-plots of representative miRNA targets and expression analysis of target genes identified from degradome sequencing. a and b The T-plots of SMARCA3L3 and CAF1 targeted by tae-miR2275-3p and tae-miR1127a. The T-plots showed the distribution of the degradome tags from the target gene sequence. miRNA cleavage was represented by red lines with spots. Cleavage sites for SMARCA3L3 and CAF1 were at 86 nt and 3820 nt, respectively. c The relative expression of selected targets from degradome data for several key miRNAs, CAF1 of tae-miR2275-3p, SMARCA3L3 of tae-miR1127a, XBP2 and EME1 of tae-miR1122c-3p, CYCA3;2 of tae-miR1122b-3p, MTHFR of tae-miR1122a, RPL23 of tae-miR9675-3p and DME of tae-miR9679–5p. The transcript level of each gene was normalized to NN1. The fold change normalized ≥ 2 or ≤ 0.5 were represented by numerical value. Error bars indicated s.d. based on three biological replicates (**P < 0.01, Student’s t-test)
