Fig. 5.
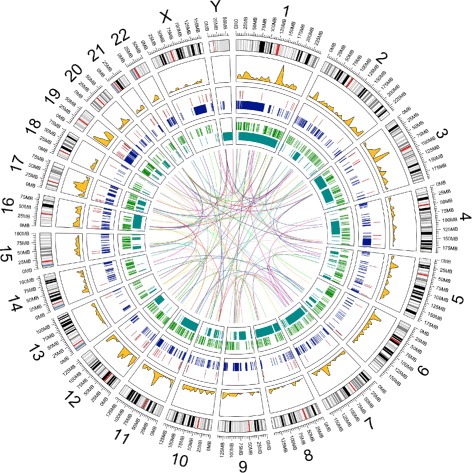
Genomic variation distributions. Distributions of multiple types of variants across the Jurkat genome. Plotted data listed from outside-in: 1. hg19 genome ideogram (gray); 2. Density of SnpEff “High Impact” SNVs with rare ExAC allele frequencies (gold); 3. Homozygous deletions that lie in coding exons (red); 4. Deletions longer than 25 kb (blue); 5. Insertions longer than 50 bp that lie in coding exons (green); 6. Inversions longer than 25 kb (cyan); 7. Interchromosomal translocations (center)
