Table 4.
Comparison of GWAS findings for DME and PDR with a previously published GWAS for sight-threatening DR in a Caucasian population (Grassi et al., 2011 [14])
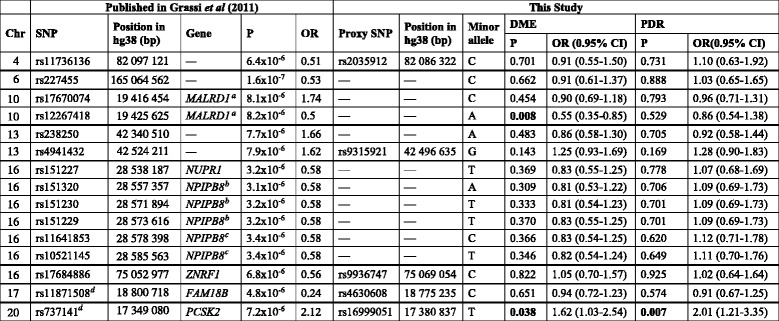
p < 0.006 was required for significance to account for the eight loci tested (as separated by horizontal bold lines). Nominally significant p-values (p < 0.05) are shown in bold. Proxy SNPs were used in this study when published SNPs were not available. Proxy SNPs were selected based on r2 value with the reported SNP in the CEU population of HapMap. 95% CI for the OR was not reported by Grassi et al., 2011 [14])
aIn Grassi et al. 2011, the MALRD1 locus was labelled as C10orf112 and NPIPB8 was labelled as CCDC101b and SULT1Ac
dSNP with lowest p-value within 50 kb of the published SNP (no appropriate proxy SNP identified)
