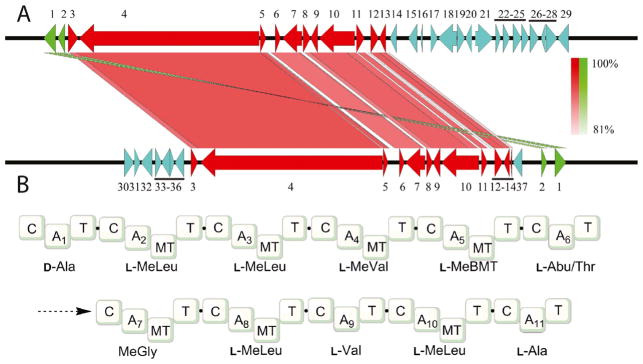
Fig. 5.
a Comparison of gene clusters responsible for biosynthesis of cyclosporin A from T. inflatum NRRL 8044 (lower) and cyclosporin C from A. felina TTI-0347 (NRRL 66746) (upper). Numbered genes in are orthologs. Intensity of shading between orthologs is proportional to identity of DNA sequences. Genes in blue indicate shared gene orthologs not involved in cyclosporin biosynthesis. Genes in green are transposed. Genes numbered 1–37 are: 1, hypothetical protein; 2, bZIP transcription factor; 3, F-box domain protein; 4, cyclosporin synthetase; 5, alanine racemase; 6, cyclophilin; 7, ABC transporter; 8, esterase-like protein; 9, cytochrome b2-like protein; 10, PKS; 11, hypothetical protein; 12, cytochrome P450; 13, aminotransferase; 14, hypothetical protein; 15–29, hypothetical proteins; 30, phosphotransferase family; 31, hypothetical protein; 32, ribonuclease P complex subunit (Pop1); 33, hypothetical protein; 34, Hsp70-like protein; 35, hypothetical protein; 36, C2H2 Zn-finger transcription factor; 37, dehydrogenase. b Predicted NRPS domain architecture and adenylation domain selectivity for the 11-amino-acid-encoding cyclosporin synthetase enzyme (gene 4). Abbreviations: C, condensation domain; A, adenylation domain; T, peptidyl carrier domain; MT, methyltransferase domain; BMT, butenyl-methyl-L-threonine; Abu, 2-aminobutyric acid