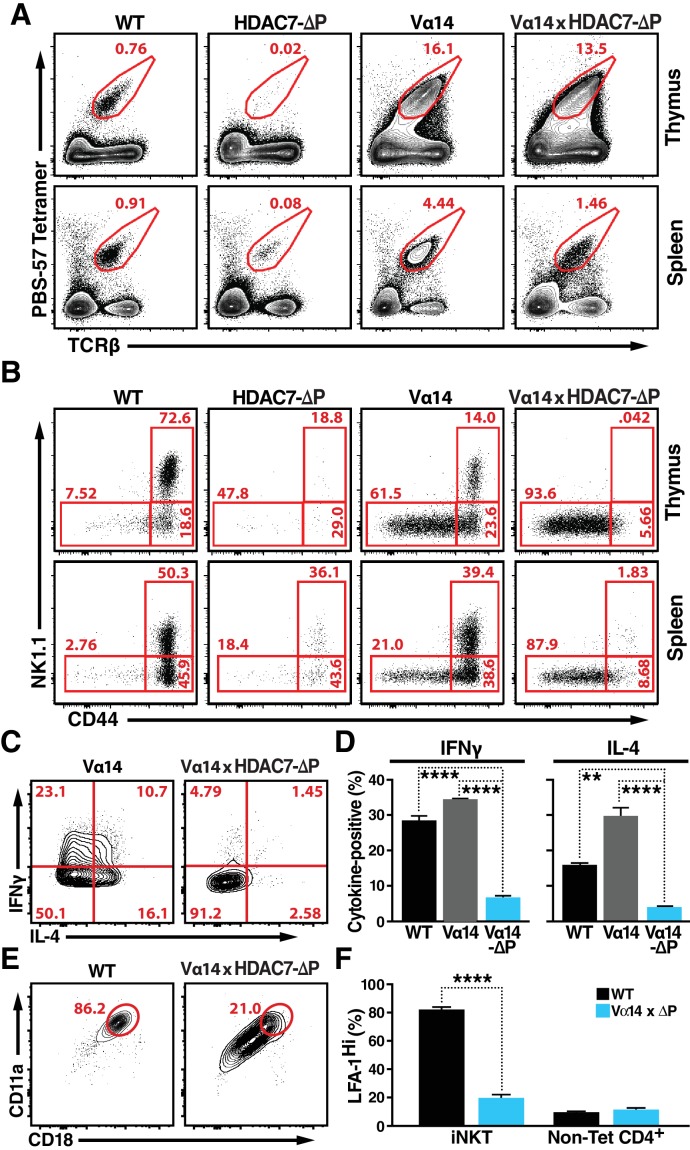
Figure 3. HDAC7-ΔP Blocks Innate Effector Development in iNKT Cells and Converts Them to Naive-Like T-cells.
(A, B) Representative flow cytometric plots showing TCRβ vs. PBS-57 tetramer staining (A), and CD44 vs. NK1.1 staining of iNKT cells (B) in thymus (top) and spleen (bottom) of littermate mice with the indicated genotypes. (C, D) Representative staining (C) and total quantification (D) of IFNγ and IL-4 secretion in total splenocytes from littermate mice of the indicated genotypes, stimulated ex vivo for 4 hr with PMA/Ionomycin. (E, F) Representative flow plots (E) showing surface expression of LFA-1 (CD11a/CD18) in splenic iNKT (Tet+/TCRβ+) cells, with quantification for four littermate pairs shown in (F). Bars on graphs indicate mean ±SEM. Data in (D, F) are from three independent experiments. Statistical significance was determined using one-way (E) or two-way (F) ANOVA; *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001. Tukey (E) or Bonferroni post-tests (F) were used for pairwise comparisons.