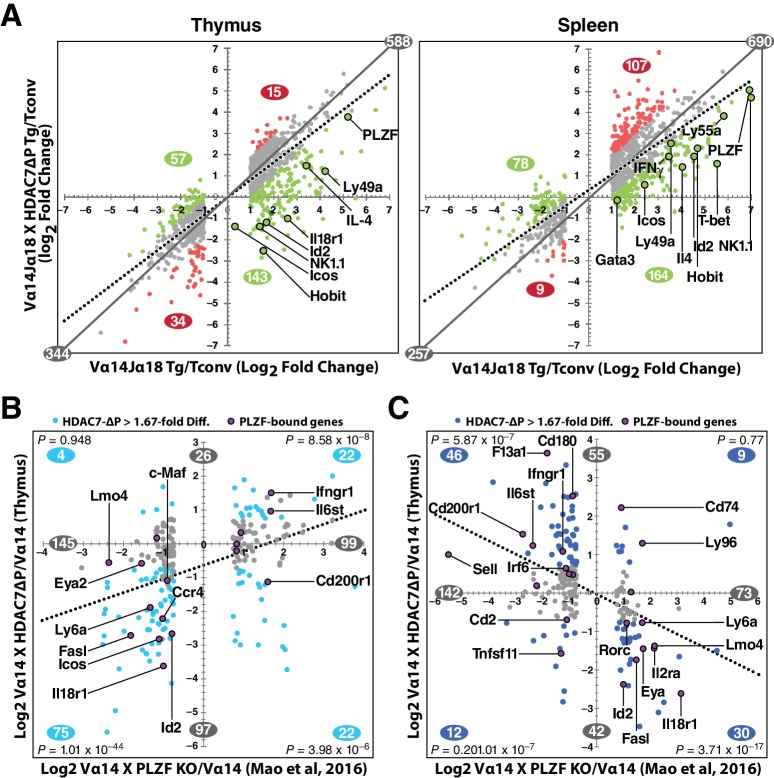
Figure 5. HDAC7 Regulates a Cassette of Genes in Glycolipid-Reactive Cells That is Highly Relevant to Innate Effector Function, Inflammation, Autoimmunity, and Autoimmune Liver Disease.
(A) Scatter charts showing gene expression changes in Cd1d/αGalCer-reactive Vα14 Tg (X axis) and Vα14 X HDAC7-ΔP Tg (Y axis) thymocytes (left) or CD4 splenocytes (right) vs naïve CD4SP thymocytes or splenocytes, respectively. The solid gray line indicates the plot diagonal and the dotted gray line indicates the Least Squares best-fit line of the plotted data. Genes displayed were expressed at least 1.75-fold differentially between tetramer-reactive and naïve cells, with p<0.05 (2-tailed Student’s T test) for three biological replicates of each genotype. Colored plot points represent genes whose differential expression vs. naïve was enhanced (red points) or suppressed (green points) at least 1.75-fold by co-expression of HDAC7-ΔP (C, D) Scatter charts showing genes > 1.66 fold differentially expressed due to loss of PLZF function (C, horizontal axis), or due to transgenic expression of PLZF (D, horizontal axis) according to (Mao et al., 2016), plotted against effect of HDAC7-ΔP expression in PBS-57 tetramer-reactive Vα14 X HDAC7-ΔP vs Vα14 Transgenic thymocytes in Thymus (C, vertical axis) or spleen (D, vertical axis). Total number of genes > 1.67 fold differentially expressed along each axis are indicated in gray. Numbers of genes, with P-values (binomial distribution) of the overlap, for genes differentially expressed along both axes in each quadrant (blue symbols), are indicated in blue.