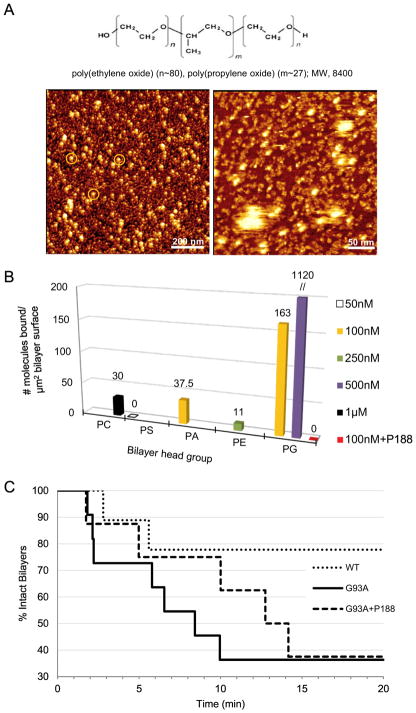
Figure 6.
P188 suppresses G93ASOD1 membrane toxicity. (A, upper) Chemical structure of tri-block copolymer P188. (A, lower) P188 molecules visualized directly using HRF-AFM. (Left image) Micelles each show a compacted hydrophobic core of ~20 nm in diameter (white spots) with a surrounding hydrophilic corona with a halo of ~5 nm (orange circles, left image). (Right image) Close inspection of a smaller area scan reveals a sub-monolayer coating of lumpy 10 nm aggregates, some assembled into higher ordered 10 nm wide twisted worm-like structures (right image) situated between the ~20 nm particles seen in A. P188 was adsorbed from a solution of 1 mM P188 in water and then scanned at RT. Full scale z-axis, 10 nm. (B) Binding assays as measured by HRF-AFM. Solutions of G93ASOD1 were incubated against each of 5 planar lipid bilayers composed of differing head groups. Binding of G93ASOD1 to PG is seen, but relatively little to PC, PS, PA or PE membranes. When 10 μM P188 is added to the assay, G93ASOD1 is no longer able to bind PG membrane (red square). Multiple areas across the bilayer surfaces were sampled to confirm representative binding. WTSOD1, the control Src homology 3 (SH3) domain of c-Src kinase, and avidin proteins showed little to no binding against the bilayers. (C) Single bilayer electrophysiology (SB-EP) shows that P188 suppresses PG-selective G93ASOD1 membrane toxicity. After ~10 min, G93ASOD1 reached its maximum toxicity on the PG-enriched reporting membrane (7 of 11 reporting membranes were ruptured, solid line). In contrast, membrane rupture was suppressed by a factor of ~2 (only 3 of 11 reporting membranes were ruptured, dashed line) in the presence of P188. In separate control experiments, 100% of the PG:PE reporting membranes that were stable for 2–3 min after formation maintained their stability over a total of ~20 min in buffer or P188 solution.