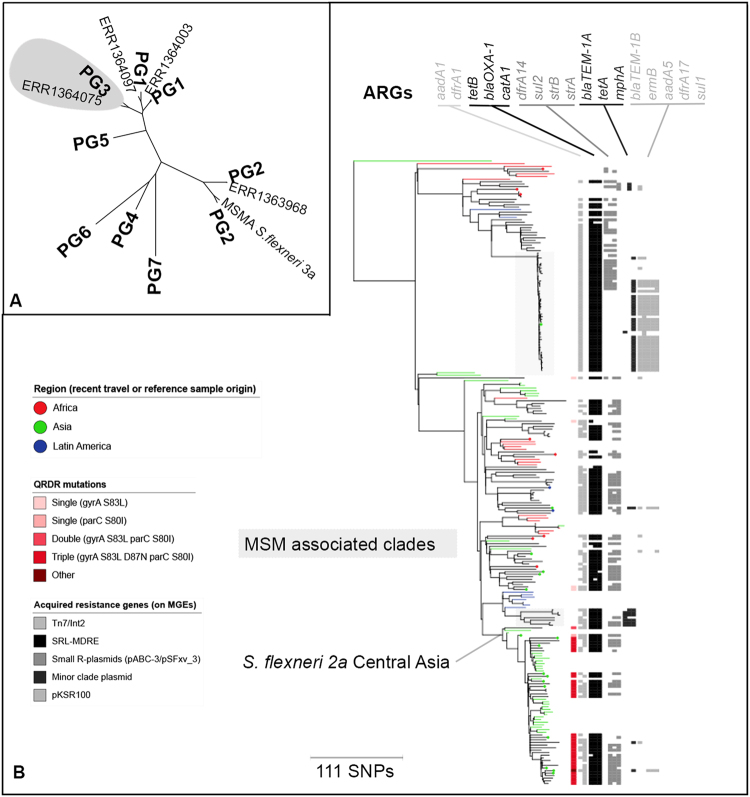
Figure 2.
Shigella flexneri 2a UK cross section in context. (A) The smaller tree shows the broad context of phylogroups (PG) 1–7 of S. flexneri and intercontinental MSM-associated (MSMA) S. flexneri 3a, with the relationships of three non-PG3 isolates from this study. PG3 is overlaid by a grey polygon in the smaller tree and is shown in full in the larger tree. (B) The larger tree is a mid-point rooted maximum likelihood tree and shows the detailed evolutionary relationships of UK cross section isolates (black branches) with reference isolates (coloured branches). Reference isolate branches are coloured by region of sample origin, and region of recent travel is overlaid for UK samples as coloured circles on tree tips (both coloured according to the inlaid region key). Two MSM-associated sublineages are highlighted by grey boxes, and the S. flexneri 2a Central Asia lineage is indicated at the defining internal node. AMR features are shown in adjacent tracks. The leftmost track shows the presence of QRDR mutations and one ‘Other’, which designates a quadruple mutant also carrying the qnrS1 gene. Subsequent gray scale tracks indicate the presence of antimicrobial resistance genes (ARGs, labelled above) associated with known Shigella MGEs. Specifically (from left to right), the Tn7/Int2, SRL-MDRE, Small R-plasmids, Minor clade plasmid, pKSR100 (Table 1).