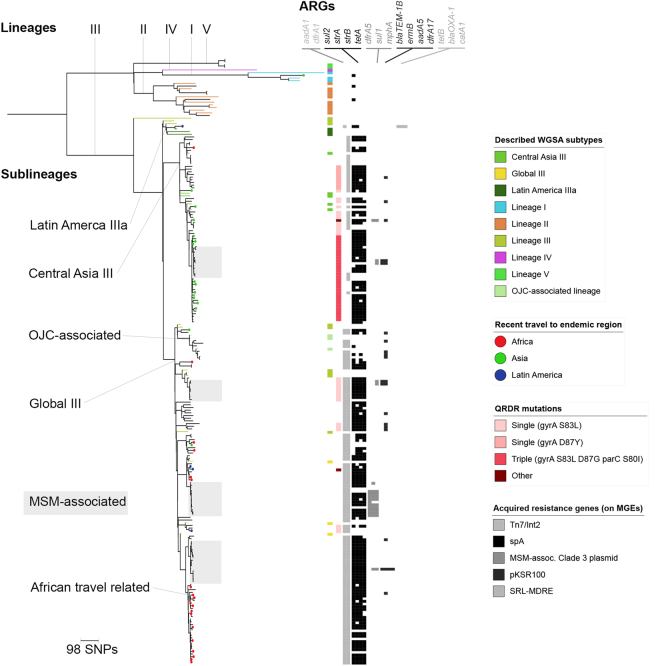
Figure 3.
Shigella sonnei UK cross section in context. The midpoint-rooted maximum likelihood phylogenetic tree shows isolates from this study in the context of five global lineages of S. sonnei. Lineage III has further relevant sublineages indicated at the internal nodes and MSM-associated sublineages are highlighted in grey. Isolates from patients recently returned from endemic regions are indicated by circles overlying the tree tips which are coloured according to the inlaid key. Reference isolates from previously described WGSA subtypes (lineages/sublineages) are indicated in the leftmost adjacent track, and tree branches for references isolates are also coloured according to the inlaid key. AMR features are shown in subsequent tracks. The second leftmost track shows the presence of QRDR mutations and two isolates containing ‘Other’ quinolone resistance determinants (including an isolate with a single QRDR mutation and a qepA gene, and an isolate containing a qnrB19 gene). Subsequent gray scale tracks indicate the presence of antimicrobial resistance genes (ARGs, labelled above) associated with known Shigella MGEs. Specifically (from left to right), the Tn7/Int2, SpA, MSM-associated sublineage 3 plasmid, pKSR100, SRL-MDRE (Table 1).