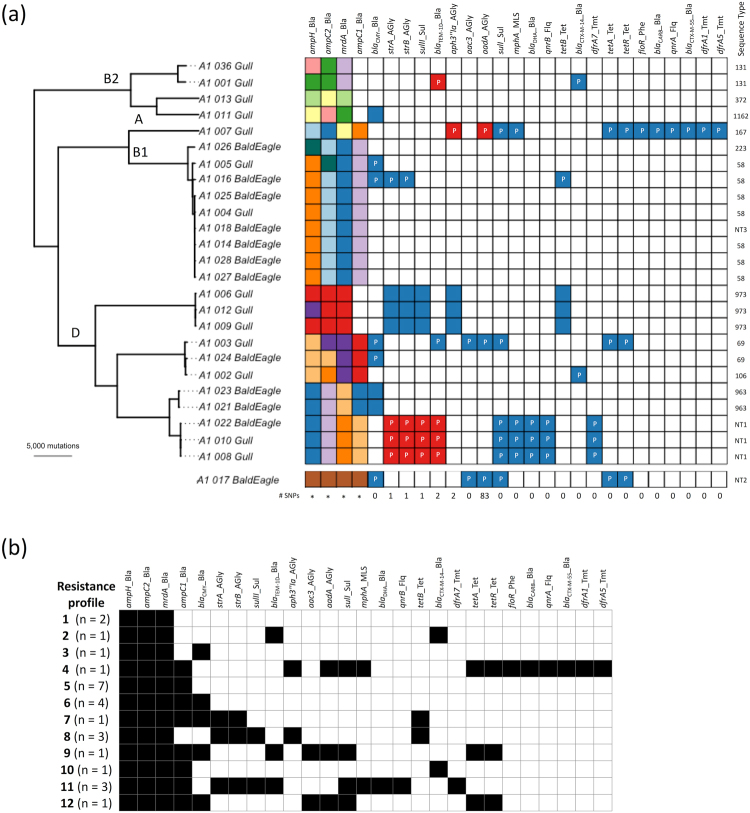
Figure 2.
(a) Midpoint rooted clonal core genome phylogeny of 25 cephalosporin-resistant E. coli isolates originating from gulls and bald eagles in Alaska. Phylotypes (A, B1, B2, D) are indicated on branches. Presence of each of 27 identified AMR genes is shown as a matrix, with gene names indicated at the top of the matrix underscored by the antibiotics class (AGly: aminoglycosides, Bla: beta-lactamases, Flq: fluoroquinolones, MLS: macrolide-lincosamide-streptogramin, Phe: phenicols, Sul: sulfonamides, Tet: tetracyclines and Tmt: trimethoprim). Coloured squares indicate presence and white indicates that the gene was not found. Each colour indicates a different allele for each gene. The number of SNPs differentiating AMR genes with two alleles is shown below the matrix, while the phylogenetic diversity of genes with more than two alleles (indicated by *) can be seen in Fig. 3. AMR genes found on plasmids identified by PlasmidSPAdes are indicated with the letter “P”. The final column of the matrix indicates in silico identified E. coli sequence types. The divergent A1_017_BaldEagle isolate is included at the bottom of the matrix, as it was omitted from the midpoint rooted clonal core genome phylogeny. (b) Unique resistance profiles identified through in silico AMR gene detection in cephalosporin-resistant E. coli isolates. A presence/absence matrix of 27 identified AMR genes is shown, with black shading indicating presence, and no shading indicating that the gene was not found. The number of isolates found with each resistance profile is indicated in parentheses.