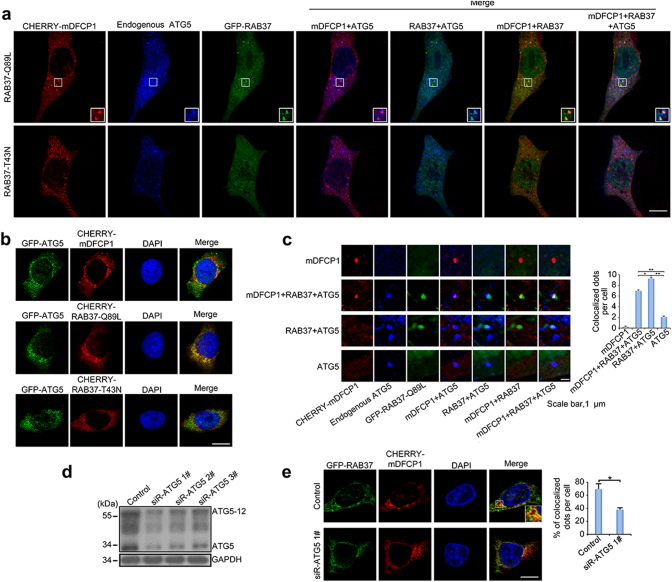
Fig. 6.
RAB37 puncta are co-localized with isolation membranes. a Co-localization of RAB37 puncta with ATG5 and an omegasome marker DFCP1. COS-7 cells were co-transfected with RAB37 mutants (GFP-RAB37-Q89L and -RAB37-T43N) and CHERRY-mDFCP1. The cells were starved in the medium EBSS for 1 h befor harvest. Endogenous ATG5 protein was stained by immune fluorescence using anti-ATG5 antibody and then anti-rabbit IgG AMCA conjugated antibody (blue). Confocal images were taken. Scale bar, 10 µm. The outlined region is magnified in the inset. b ATG5 co-localizations with RAB37-Q89L, RAB37-T43N and DFCP1, respectively. GFP-ATG5 was co-transfected with CHERRY-RAB37-Q89L, CHERRY-RAB37-T43N and CHERRY-DFCP1 in HeLa cells, respectively. Scale bar, 10 µm. c Representative images of DFCP1-positive, RAB37-Q89L/DFCP1/ATG5-positive, RAB37-Q89L/ATG5-positive and ATG5-positive puncta in COS-7 cells from (a). Positive dots were quantified from ~20 cells. Scale bar, 1 µm. Data are presented as means ± S.D. * stands for P < 0.05, ** stands for P < 0.01 (n = 3 independent experiments). d Determination of effective ATG5 siRNA. Three siRNAs and a negative control siRNA were transfected into HeLa cells, and ATG5 and ATG5-12 protein levels were analysed by Western blots. e ATG5 interference inhibits RAB37 co-localization with CHERRY-mDFCP1. HeLa cells were co-transfected with siRNA (siR-ATG5 1# or negative control), GFP-RAB37 and CHERRY-mDFCP1. The cells were cultured in DMEM with 10% FBS and starved in starvation medium EBSS for 1 h. The images were taken under confocal microscopy. Scale bar, 10 µm. Percentage of co-localized dots (GFP-RAB37 + CHERRY-mDFCP1 (yellow) / CHERRY-mDFCP1 (red)) in controls were determined in comparison with ATG5 knockdown treatments. The co-localized dots were counted from ~20 cells. * stands for P < 0.05