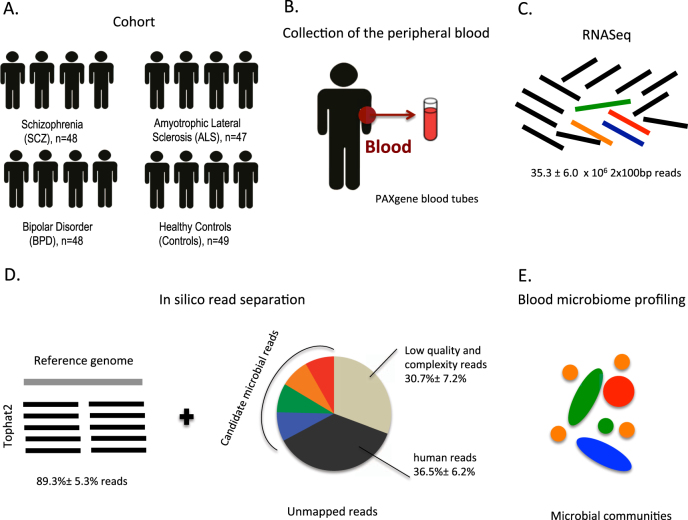
Fig. 1. Microbial profiling using RNA-Seq data from whole blood.
a We analyzed a cohort of 192 individuals from four subject groups, i.e. Schizophrenia (SCZ, n = 48), amyotrophic lateral sclerosis (ALS n = 47), bipolar disorder (BPD n = 48), unaffected control subjects (Controls n = 49). b Peripheral blood was collected for RNA collection. c RNA-Seq libraries were prepared from total RNA using ribo-depletion protocol. d Reads that failed to map to the human reference genome and transcriptome were sub-sampled and further filtered to exclude low-quality, low complexity, and remaining potentially human reads. e High quality, unique, non-host reads were used to determine the taxonomic composition and diversity of the detected microbiome. See also Table S1