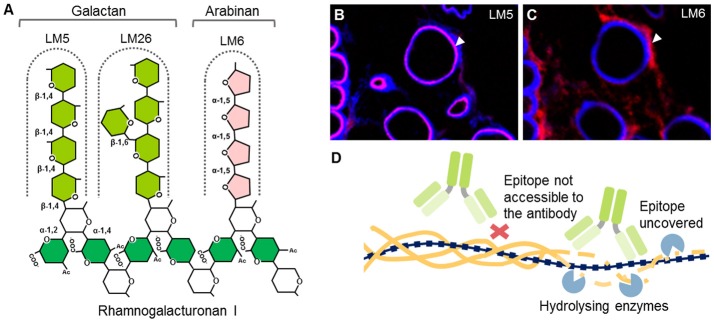
Figure 3.
Example of mAb-recognized epitopes, cell wall heterogeneity, and masking. (A) Example of different epitopes recognized by mAbs on RG-I side chains. Both LM5 and LM26 mAbs bind to (1 → 4)-β-D galactan epitopes with a difference in requirement of a Gal substitution via α-D-(1 → 6) bond in the case of LM26. LM6 requires a linear chain of four α-(1 → 5) linked L-arabinose units. This epitope can be found also on AGPs. (A,B) LM5 and LM6 mAbs can have affinity toward different cell wall microdomains. An example of an in situ labeling of sections of resin embedded pea border cells with LM5 and LM6. Calcofluor White (blue channel) and signal from the secondary anti-rat antibody conjugated to Alexa Fluor 555 (red). (B) LM5 labels cell walls of released border cells, whereas (C) LM6 labels the shed cell wall material released to the environment (arrowheads). For the original experiments see Mravec et al. (2017a). (D) Phenomenon of masking of mAbs epitopes. Cell wall carbohydrates are arranged in tight arrays and this could prevent binding of mAbs. Pre-digestion with specific enzymes can reveal these “hidden” epitopes.