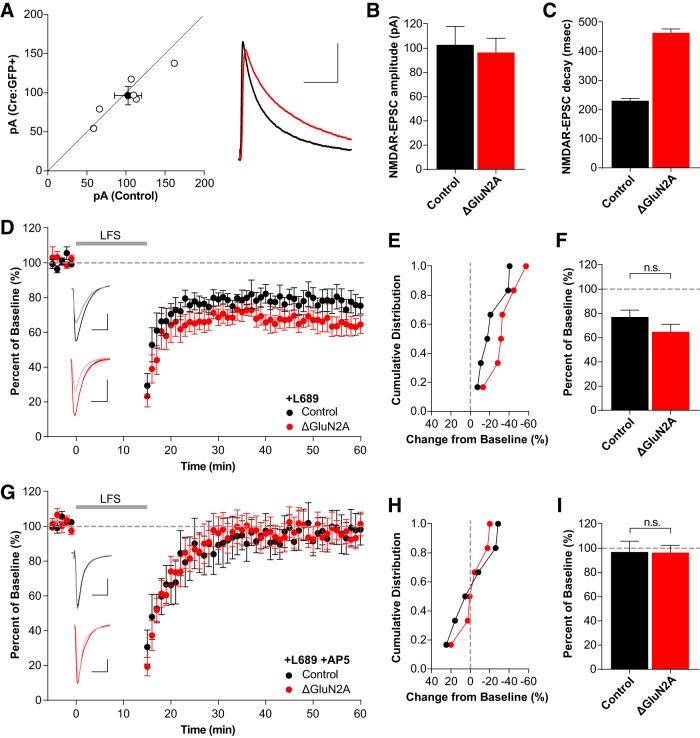
Figure 3.
Single-neuron deletion of GluN2A does not prevent non-ionotropic LTD. A–C, Single-neuron deletion of GluN2A. A, Scatterplot of individual neuron pairs (open circles) and averaged pair ± SEM (solid circle). Sample trace scale bars indicate 100 ms, 40 pA. B, Average NMDAR-EPSC amplitudes for control (102.8 ± 15.2 pA, n = 6) and Cre:GFP+ neurons (96.4 ± 11.9 pA, n = 6); p = 0.48. C, GluN2A deletion results in significantly longer decay kinetics (control: 230.4 ± 8.5 ms, Cre:GFP+: 414.4 ± 13.3 ms; p < 0.0001). D–F, GluN2A deletion does not block LTD. D, Averaged whole-cell LTD experiments and representative traces (10 ms, 50 pA). E, Cumulative distribution of experiments in D. F, Average percentage depression relative to baseline; control neurons (77.1 ± 5.8%, n = 6), Cre:GFP+ neurons (ΔGluN2A: 65.1 ± 6.2%, n = 6; t(10) = 1.431, p = 0.183, t test) G–I, LTD after GluN2A deletion is still blocked by AP5. G, Averaged whole-cell LTD experiments and representative traces (10 ms, 50 pA). H, Cumulative distribution of experiments in G. I, Summary graph of average percentage depression relative to baseline; control neurons (97.0 ± 9.0%, n = 6), Cre:GFP+ neurons (ΔGluN2A: 96.7 ± 6.0%, n = 6; t(10) = 0.0274, p = 0.979, t test).