Abstract
Objective
Regulation of intercellular adhesion molecule (ICAM)-1 in retinal endothelial cells is a promising druggable target for retinal vascular diseases. The ICAM-1-related (ICR) long non-coding RNA stabilizes ICAM-1 transcript, increasing protein expression. However, studies of ICR involvement in disease have been limited as the promoter is uncharacterized. To address this issue, we undertook a comprehensive in silico analysis of the human ICR gene promoter region.
Results
We used genomic evolutionary rate profiling to identify a 115 base pair (bp) sequence within 500 bp upstream of the transcription start site of the annotated human ICR gene that was conserved across 25 eutherian genomes. A second constrained sequence upstream of the orthologous mouse gene (68 bp; conserved across 27 Eutherian genomes including human) was also discovered. Searching these elements identified 33 matrices predictive of binding sites for transcription factors known to be responsive to a broad range of pathological stimuli, including hypoxia, and metabolic and inflammatory proteins. Five phenotype-associated single nucleotide polymorphisms (SNPs) in the immediate vicinity of these elements included four SNPs (i.e. rs2569693, rs281439, rs281440 and rs11575074) predicted to impact binding motifs of transcription factors, and thus the expression of ICR and ICAM-1 genes, with potential to influence disease susceptibility. We verified that human retinal endothelial cells expressed ICR, and observed induction of expression by tumor necrosis factor-α.
Electronic supplementary material
The online version of this article (10.1186/s13104-018-3384-8) contains supplementary material, which is available to authorized users.
Keywords: Intercellular adhesion molecule, ICAM-1, ICAM-1-related, Promoter
Introduction
Background
Retinal vascular diseases occur commonly and represent a major cause of blindness in developing and developed nations. These conditions include diabetic retinopathy, major retinal vessel occlusions, sickle cell retinopathy, retinopathy of prematurity and uveitis [1]. A key feature of the pathology of any retinal vasculopathy is over-expression of endothelial adhesion molecules in response to stimuli such as inflammatory molecules, toxic metabolites and hypoxia. The increased expression of adhesion molecules promotes leukocyte interactions with the endothelium, resulting in leukostasis, migration of leukocytes into the tissue and/or endothelial cell apoptosis [1]. Present evidence [2–6] implicates intercellular adhesion molecule (ICAM)-1, which is expressed at relatively high levels on human retinal endothelium [7], as a critical player.
Targeting retinal endothelial ICAM-1 for therapeutic purposes has been entertained [1]. However, ICAM-1 plays an important role in host immune defense and complete blockade would pose the risk of infection. In previously published work [8], we showed that a stimulus-induced increase in ICAM-1, but not its basal level, might be reduced by manipulating gene expression. Small interfering RNA knock-down of the multi-functional transcription factor (TF), nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB)1, in human retinal endothelial cells significantly reduced a tumor necrosis factor (TNF)-α-induced increase in ICAM-1 expression, but did not alter constitutive ICAM-1 expression. Importantly, NF-κB1 knock-down also significantly reduced leukocyte binding to TNF-α-stimulated human retinal endothelial cell monolayers, but did not impact baseline binding.
Rationale
Specificity is an important consideration for success in planning a treatment approach that involves manipulating ICAM-1 expression. One candidate molecule with a primary function in regulating ICAM-1 expression is ICAM-1-related (ICR), which is a long non-coding (lnc)RNA transcribed from the anti-sense DNA strand overlapping the ICAM1-ICAM4-ICAM5 gene cluster on chromosome 19p13.2. This lncRNA was described recently in a publication by Guo et al. [9], who studied a cell line generated from portal vein thrombus of an individual with hepatocellular carcinoma [10]; the investigators showed ICR bound to and stabilized the ICAM-1 transcript, leading to increased ICAM-1 protein expression. Prior to considering ICR blockade as a treatment of retinal vasculopathy, it is essential to understand how disease-relevant stimuli trigger transcription of ICR. To date, however, the ICR promoter has not been characterized. Thus, we undertook a comprehensive in silico analysis of ICR promoter to identify potential TF binding sites (TFBSs), as well as associations between TFBSs and single nucleotide polymorphisms (SNPs). We also verified that human retinal endothelial cells expressed ICR.
Main text
Functionally constrained elements in ICR gene promoter regions
Conserved sequences across orthologous promoters may identify TFBSs of functional relevance; conserved sequences signify genomic regions that have resisted evolutionary mutation over time, implying a functional constraint [11]. Sequences for the human ICR gene and an orthologous mouse gene have been manually annotated by the HAVANA (Human and Vertebrate Analysis and Annotation) project and lodged with Ensembl [12] (human: AC011511.5 and ENSG00000267607.1; mouse: AC159314.1 and ENSMUSG00000110790). The human gene consists of a single exon that begins in the ICAM4-ICAM5 intergenic region, spans the entire ICAM4 gene, and overlaps the 3′ untranslated region (UTR) of ICAM1 (Fig. 1). The mouse gene, which is located on chromosome 9, also spans Icam4 and overlaps the 3′ UTR of Icam1, but has 2 exons: the first exon begins within intron 1 of Icam5 and continues across exon 1 of Icam5, and the second exon begins in the Icam4-Icam5 intergenic region and spans Icam4 (Fig. 1). Since locations of the transcription start site (TSS) and, by extrapolation, the promoter region, differ between human and mouse genes, we conducted separate analyses to identify regions of constrained DNA sequence across multiple eutherians.
Fig. 1.
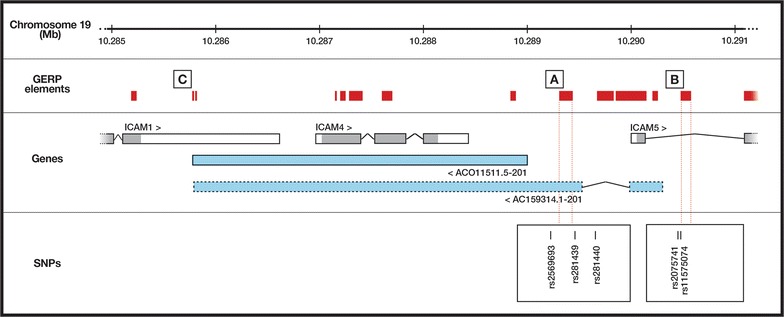
Genomic structure of human ICAM-1-related (ICR) long non-coding RNA. Positions of annotated human ICR gene transcript (AC011511.5-201, solid line: ENST00000589379.1) and predicted alternative transcript (based on annotated mouse sequence: AC159314.1-201, dashed line: ENSMUST00000216917.1), both located on the anti-sense strand, are indicated in relation to positions of ICAM1 (ENST00000264832.7), ICAM4 (ENST00000380770.3) and ICAM5 (ENST00000221980.4) on chromosome 19p13.2 (genome build = GRCh38.p10; grey indicates coding sequence). Genome evolutionary rate profiling (GERP) identifies evolutionarily constrained elements within a region 500 base pairs upstream of the transcription start sites of the human (A: conserved across 25 eutherians) or mouse (B: conserved across 27 eutherians) ICR gene sequences, plus two polyadenylation signals at the 3′ end of both ICR gene sequences (C: conserved across 26 or 24 eutherians). Phenotype-associated single nucleotide polymorphisms (SNPs) predicted to impact binding of factors that may regulate ICR gene transcription are showed in boxes that correspond to GERP-defined elements
Genomic evolutionary rate profiling (GERP) was performed using the Ensembl genome browser (Ensembl release 89—May 2017) [12] to identify evolutionarily constrained elements across annotated eutherian sequences located within the region 500 base pairs (bp) upstream of the human TSS and the mouse TSS [13]. The human genome build was GRCh38.p10, and the mouse genome build was GRCm38.p5. Genome sources for other eutherians are listed in Additional file 1: Table S1. One GERP-constrained element of 115 bp (located at − 276 to − 390 bp relative to the TSS) in the human ICR promoter was common to 25 genomes; the dolphin sequence was excluded, as the match was incomplete across the region. A second GERP-constrained element of 68 bp (located at − 177 to − 244 bp relative to the TSS) in the promoter of the orthologous mouse gene located on chromosome 9 was conserved across 27 genomes; the pika and lesser hedgehog tenrec sequences were similarly excluded due to incomplete matching. The proximity of these conserved, non-coding DNA sequences to the human or mouse TSS indicate a likely role in regulating ICR gene expression. Interestingly, the GERP-constrained element in the mouse gene is conserved in the human genome, suggesting a second ICR transcript—with two exons—may exist in the human. Two polyadenylation signals at the 3′ end of the ICR gene are also conserved across 26 and 24 eutherian genomes, including human and mouse.
Transcription factor binding sites within conserved ICR gene promoter regions
The GERP-constrained elements—plus 10 bp upstream sequence and 10 bp downstream sequence—within the defined promoter regions of the human ICR and orthologous mouse genes were interrogated for predicted TFBSs in the ‘General Core Promoter Elements’ and ‘Vertebrates’ sections of Matrix Library 10.0 (October 2016) using MatInspector [14] in Genomatix Software Suite (http://www.genomatix.de): the ‘Common TFs’ search tool was used; ‘Core similarity’ was set at 0.75; and ‘Matrix similarity’ was set at optimized, to minimize false positives for individual matrices. Searching the human element identified 5 matrix families (containing 15 matrices) common to all 24 eutherian sequences, plus one additional family (containing 2 matrices) in 23 of the 24 sequences. These matrices predicted TFBSs for gene regulators including: HIF1, NRF1, MYC-MAX, BHLHE40, XBP1 and STAF. Searching the mouse element identified one matrix family (containing 8 matrices) in 24 of 25 eutherian sequences, a second family (containing 5 matrices) in 23 of 25 sequences, and a third family (containing 3 matrices) in 19 of 25 sequences. These matrices predicted TFBSs for DNA-binding factors that include: RFX1, AP-1 and BACH2. Matrix families, matrices and predicted binding sites are listed in Table 1, and sequences and locations corresponding to each matrix are provided in Additional file 2: Table S2. The spectrum of TFBSs assigned to the ICR gene promoter in our analysis is consistent with transcription in response to a wide range of pathological stimuli that include hypoxia, and metabolic and inflammatory proteins.
Table 1.
Predicted transcription factor binding sites in evolutionarily conserved elements, as identified by genomic evolutionary rate profiling of the sequence located 500 bp upstream of human ICAM-1-related gene transcription start sites
| Human sequence | Matrix familya | Matrixa | Description |
|---|---|---|---|
| Annotated | V$HIFF | V$ARNT.01 | AhR nuclear translocator homodimers |
| V$HIF1.02 | Hypoxia inducible factor | ||
| V$CLOCK_BMAL1.01 | Clock/BMAL1, NPAS2/BMAL1 heterodimers | ||
| V$HRE.02 | Hypoxia-response elements | ||
| V$HRE.03 | Hypoxia response elements, HIF1a/ARNT heterodimers | ||
| V$HASF | V$HAS.01 | HIF-1 ancillary sequence | |
| V$NRF1 | V$NRF1.01 | Nuclear respiratory factor | |
| V$NRF1.02 | Nuclear respiratory factor | ||
| V$EBOX | V$MNT.01 | MAX binding protein | |
| V$MYCMAX.02 | MYC-MAX binding sites | ||
| V$MYCMAX.03 | MYC-MAX binding sites | ||
| V$NMYC.01 | N-myc | ||
| V$USF.04 | Upstream stimulating factor ½ | ||
| V$HESF | V$DEC1.02 | Basic helix-loop-helix protein E40 (BHLHE40) | |
| V$HELT.01 | Hey-like transcriptional repressor | ||
| V$CREB | V$XBP1.01 | X-box binding protein 1 | |
| V$TAXCREB.02 | Tax/CREB complex | ||
| V$STAF | V$STAF.01 | Se-Cys tRNA gene transcription activating factor | |
| Predicted (based on mouse) | V$XBBF | V$RFX1.02 | X-box binding protein RFX1 |
| V$RFX2.02 | Regulatory factor X, 2 | ||
| V$RFX3.01 | Regulatory factor X, 3 | ||
| V$RFX4.01 | Regulatory factor X, 4 | ||
| V$RFX4.03 | Regulatory factor X, 4 | ||
| V$RFX5.02 | Regulatory factor X, 5 | ||
| V$XBOX.01 | Motif bound by RFX proteins | ||
| V$MIF1.01 | Myc Intron-binding protein (MIBP)/RFX complex | ||
| V$AP1F | V$AP1.01 | Activator protein 1 | |
| V$AP1.02 | Activator protein 1 | ||
| V$BATF.01 | Basic leucine zipper TF | ||
| V$FOSL1.01 | Fos-like antigen 1 | ||
| V$JUNB.01 | Jun-B | ||
| V$AP1R | V$BACH2.02 | BTB and CNC homology 1, basic leucine zipper TF 2 | |
| V$MARE_ARE.01 | Antioxidant response elements | ||
| V$NFE2.01 | NF-E2 p45 |
aMatrix nomenclature is defined by MatInspector [7]. Matrices predict transcription factor binding sites; matrix families group individual matrices that have similar binding properties
Single nucleotide polymorphisms in proximity to ICR gene promoter regions
We sought to identify phenotype-associated SNPs with potential to influence transcription of the human ICR gene, searching both the ICR-ICAM5 intergenic region and 100 bp downstream of the ICR TSS. We also searched intron 1 of the ICAM5 gene, since GERP suggested the mouse lncRNA might be represented in the human. Single nucleotide polymorphisms within these regions (plus 25 bp upstream sequence and 25 bp downstream sequence) were extracted from the Single Nucleotide Polymorphism Database of the US National Center for Biotechnology Information [15]. The Genomatix Software Suite SNPInspector was used to search Matrix Library (search settings as described for MatInspector analysis) for promoter elements predicted to be impacted by these SNPs. Results were checked against chromatin immunoprecipitation-sequencing data in ENCODE (Encyclopedia of DNA Elements) [16] to confirm TF binding at SNP locations. This analysis identified 3 SNPs in the defined region between ICR and ICAM5 plus 100 bp downstream of the ICR TSS (rs2569693, rs281439 and rs281440), and 2 SNPs in intron 1 of ICAM5 (rs2075741 and rs11575074) (Fig. 1; Table 2, Additional File 3: Table S3). All these SNPs have been associated with human disease and/or serum levels of the membrane-cleaved form of ICAM-1, which is commonly referred to as soluble (s)ICAM-1.
Table 2.
Phenotype-associated single nucleotide polymorphisms (SNPs) and predicted impact of minor allele on binding sites for transcription factors within the human ICAM-1-related (ICR) gene promoter
| SNP | Alleles | Minor allele | Transcription factors with binding sites in ICR promoter | Impact of minor allele on binding sites | ||
|---|---|---|---|---|---|---|
| Frequency | Associations | Loss | Gain | |||
| rs2569693 | C/T | 0.31 | Systemic lupus erythematosus [1] sICAM-1 level [18] |
ELF1, POL2, NFKB, MAX, CMYC, E2F6, MXI1, FOXA1, CHD2, HMGN3, ZNF263 | PLAGL1, WT1, EGR2, ZKSCAN3, ZBED4, INSM1, MAZR, SALL2, SPZ1 |
MOK2 |
| rs281439 | C/G | 0.32 | Breast cancer [19] | USF1, USF2, USF3, FOXA1, SIN3AK20, POL2, NFKB, MAX, CMYC, E2F6, MXI1, POL24H8, TCF12, BHLHEAD, HMGN3, IRF1, TBP, YY1, ZNF263 | USF | E2F6, GCM1, XCPE |
| rs281440 | A/G | 0.30 | sICAM-1 level [20] | POL2, TBP, EGR1, SRF, IRF, POL24H8, GATA2, ZNF263 | KKLF, ZBTB7, LRRFIP1, E2F4 NM23, MAZR |
none |
| rs2075741 | G/C | 0.33 | Breast cancer [19] Prostate cancer [19] sICAM-1 level [20] |
POL2, TAF1, ZNF263 | None | DMTE |
| rs11575074 | G/A | 0.07 | sICAM-1 level [21] | POL2, TAF1, SUZ12, ZNF263 | NF1B | MYBL1, MEIS1A_ HOXA9, PTF1 |
sICAM-1 soluble intercellular adhesion molecule 1
The rs2569693 SNP has been associated with susceptibility to systemic lupus erythematosus across different human populations [17], and with sICAM-1 levels [18]. Analysis identifies the location of this SNP to coincide with binding sites for numerous TFs, including MYC and SCAN domain family members, and predicts the minor allele will cause loss of sites for MYC family member, MAZR, and SCAN domain family member, ZKSCAN3, plus gain of a site for MOK2. The minor allele at rs281439 has been associated with increased risk of breast cancer and progression of the cancer [19]. Multiple TFs, including USF family members, bind at this position, and the minor allele may code loss of the USF binding site, plus gain of E2F6, GCM1 and XCPE binding sites. The rs281440 minor allele has been linked to reduced levels of sICAM-1 [20]; it is predicted to cause no gains of binding motifs, but loss of several; however, ENCODE identified no TFs that bound this position, and thus conclusions on binding activity are not possible at this time. The minor allele at rs2075741 has been associated with breast and prostate cancer [19], and elevated levels of sICAM-1 [20]. Several TFs are predicted to bind at this site; no TFBS is lost with the minor allele, but a new MTE (designated DMTE in Genomatix Software Suite) binding sequence may be created. Similarly, analysis of rs11575074 minor allele, which also has been linked with elevated sICAM-1 levels [21] reveals no losses in known TFBSs, but sites for MYBL1, PTF1 and MEIS1-HOXA9 may be gained. Our findings suggest at least 4 of 5 SNPs are likely to influence TF binding to the ICR gene promoter, and thus alter the level of ICR and ICAM-1 expression.
Expression of ICR by human retinal endothelial cells
Given that our interest in ICR relates to its potential as a therapeutic target for ICAM-1 in retinal vasculopathy, we wanted to verify the expression and induction of ICR in endothelial cells isolated from human retinae. Tumor necrosis factor-α has been associated with the spectrum of retinal vasculopathies [1], and its signaling pathways involve TFs that recognize the TFBSs identified in our ICR promoter analysis [22, 23]. We generated endothelial cell isolates from four paired human retinae, using the method that we have previously described in detail [1]. Cells were cultured in MCDB-131 medium (Sigma-Aldrich, St. Louis, MO), supplemented with 10% heat-inactivated fetal bovine serum (FBS) (HyClone-GE Healthcare Life Sciences, Logan, UT) and endothelial growth factors (EGM-2 SingleQuots supplement, omitting FBS, hydrocortisone and gentamicin; Clonetics-Lonza, Walkersville, MD) at 37 °C and 5% CO2 in air, and used at passage 1 or 2. Confluent cells were treated with fresh medium alone or containing TNF-α (10 ng/mL, R&D Systems, Minneapolis, MN) for 4 h, and lysed with Buffer RLT (Qiagen, Hilden, Germany). Total RNA was extracted using the RNeasy mini kit (Qiagen), and reverse-transcribed using iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad, Hercules, CA). Quantitative real-time polymerase chain reaction was performed following standard methodology, using iQ SYBRGreen Supermix (Bio-Rad) and on the CFX96 Connect Real-Time PCR Detection System (Bio-Rad) [24]. Relative expression was calculated in the Gene Expression Analysis module of CFX Manager v3.1 (Bio-Rad), which uses the 2−ΔΔCt method [25], normalizing to two stable reference genes. Primer sequences are given in Additional file 4: Table S4. As presented in Fig. 2, we observed that retinal endothelial cell isolates from all four human retinae expressed ICR and ICAM-1. Cellular expression of ICR, as well as ICAM-1 transcript, was significantly increased (p < 0.05: Student t test, 2-tailed) following exposure to TNF-α. Both level of ICR expression and degree of induction varied between individual donors.
Fig. 2.
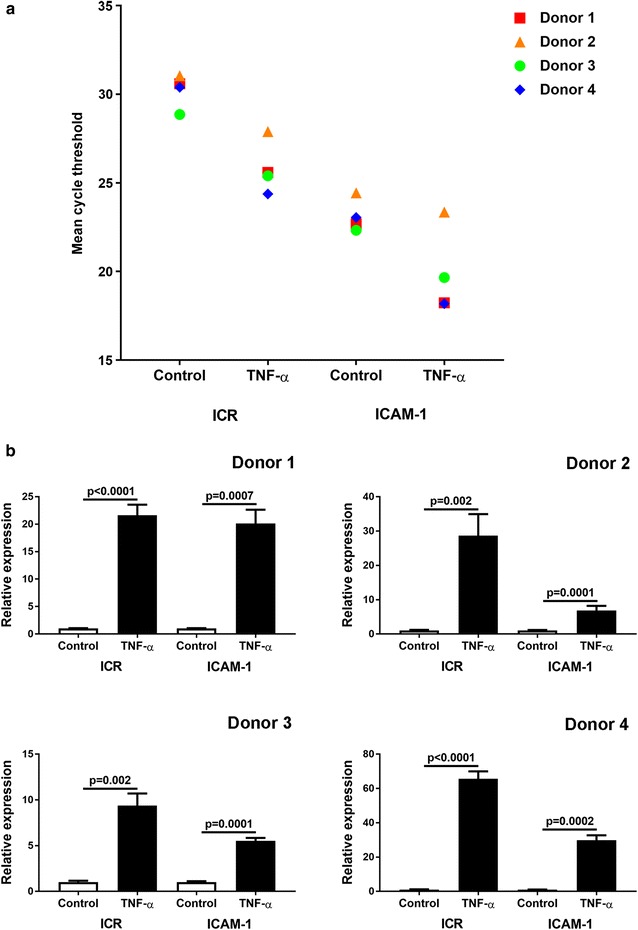
Expression of intercellular adhesion molecule (ICAM)-1-related (ICR) and ICAM-1 by human retinal endothelial cells. Primary endothelial cells were individually isolated from retinae of four different human donors, and exposed to tumor necrosis factor (TNF)-α-supplemented or control medium for 4 h (n = 3 replicate cultures/condition). a Graph shows mean cycle threshold values obtained in quantitative real-time polymerase chain reaction for ICR and ICAM-1 cDNA. Results for each human donor are represented by a unique shape, and represent (TNF)-α-supplemented versus control values. b Graphs show relative expression of ICR and ICAM-1 transcript by human donor. Bars represent mean relative expression, with error bars showing standard error of the mean. Reference genes were β-actin and glyceraldehyde-3-phosphate dehydrogenase. Data were analyzed by two-tailed Student’s t test
Limitations
Our analysis of the ICR gene promoter focused on evolutionarily conserved genomic sequences located within 500 bp upstream of the TSS. While the majority of TFBSs are expected to lie close to the TSS, it is possible that other TFBSs exist outside these regions.
This investigation was conducted in silico, and therefore it predicts candidate TFBSs, some of which may not be confirmed in biological studies. The computational analysis was comprehensive, but also limited by the capacity of present software tools and database annotations.
We verified expression and induction of ICR in human retinal endothelial cells, since our interest in this lncRNA relates to the regulation of ICAM-1 protein expression in retinal vasculopathy. Our observations may not apply to all human cell populations.
Additional files
Additional file 1: Table S1. Sources for eutherian genomes used in genome evolutionary rate profiling (GERP) and chromosomal locations of GERP-constrained elements.
Additional file 2: Table S2. Predicted transcription factor binding sites within genome evolutionary rate profiling (GERP)-constrained elements.
Additional file 3: Table S3. Description of phenotype-nucleotide polymorphisms (SNPs) predicted to influence transcription of the human ICR gene.
Additional file 4: Table S4. Primer pairs and product sizes for gene transcripts studied in human retinal endothelial cells. References are provided for primers sequences sourced from the literature.
Authors’ contributions
AL, JRS, BA conceived the study; AL, YM, LA, AS, BA generated experimental data; AL, LA, DK, JRS, BA analyzed and/or interpreted data; AL, JRS, BA drafted the paper; and DK provided critical review of the paper. All authors read and approved the manuscript.
Acknowledgements
The authors wish to thank Mr. David Heinrich (Flinders Medical Centre Medical Illustration & Media Unit, Adelaide, Australia) for his assistance with graphical design.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in the manuscript.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The use of human cadaver donor eyes from the Eye Bank of South Australia (Adelaide, Australia) was approved by the Southern Adelaide Clinical Human Research Ethics Committee (Project ID. 175.13).
Funding
This work was supported in part by: Australian Research Council (JRS: FT130101648); National Health and Medical Research Council Australia (JRS: APP1123684); the Ophthalmic Research Institute of Australia (JRS); and Flinders Faculty of Medicine, Nursing & Health Sciences (AL, DK, JRS, BA). The funders had no role in study design, in data collection, analysis and interpretation, or in writing the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Bp
base pairs
- FBS
fetal bovine serum
- GERP
genomic evolutionary rate profiling
- ICAM-1
intercellular adhesion molecule 1
- ICR
ICAM-1-related
- lncRNA
long non-coding RNA
- NF-κB1
nuclear factor κ-light-chain-enhancer of activated B cells
- SNP
single nucleotide polymorphism
- TF
transcription factor
- TFBS
transcription factor binding site
- TNF-α
tumor necrosis factor alpha
Footnotes
Justine R. Smith and Binoy Appukuttan contributed equally to this work and share senior authorship of this manuscript
Electronic supplementary material
The online version of this article (10.1186/s13104-018-3384-8) contains supplementary material, which is available to authorized users.
Contributor Information
Amanda L. Lumsden, Email: amanda.lumsden@flinders.edu.au
Yuefang Ma, Email: yuefang.ma@flinders.edu.au.
Liam M. Ashander, Email: liam.ashander@flinders.edu.au
Andrew J. Stempel, Email: Andrew.stempel@flinders.edu.au
Damien J. Keating, Email: damien.keating@flinders.edu.au
Justine R. Smith, Email: justine.smith@flinders.edu.au
Binoy Appukuttan, Email: binoy.appukuttan@flinders.edu.au.
References
- 1.Bharadwaj AS, Appukuttan B, Wilmarth PA, Pan Y, Stempel AJ, Chipps TJ, et al. Role of the retinal vascular endothelial cell in ocular disease. Prog Retin Eye Res. 2013;32:102–180. doi: 10.1016/j.preteyeres.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mcleod DS, Lefer DJ, Merges C, Lutty GA. Enhanced expression of intracellular adhesion moleule-1 and P-selectin in the diabetic human retina and choroid. Am J Pathol. 1995;147:642–653. [PMC free article] [PubMed] [Google Scholar]
- 3.Holm M, Morken TS, Fichorova RN, VanderVeen DK, Allred EN, Dammann O, et al. Systemic inflammation-associated proteins and retinopathy of prematurity in infants born before the 28th week of gestation. Invest Ophthalmol Vis Sci. 2017;58:6419–6428. doi: 10.1167/iovs.17-21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noma H, Funatsu H, Mimura T, Eguchi S. Vitreous inflammatory factors and serous retinal detachment in central retinal vein occlusion: a case control series. J Inflamm. 2011;8:38. doi: 10.1186/1476-9255-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunz Mathews M, Mcleod DS, Merges C, Cao J, Lutty GA. Neutrophils and leucocyte adhesion molecules in sickle cell retinopathy. Br J Ophthalmol. 2002;86:684–690. doi: 10.1136/bjo.86.6.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bharadwaj AS, Schewitz-Bowers LP, Wei L, Lee RW, Smith JR. Intercellular adhesion molecule 1 mediates migration of Th1 and Th17 cells across human retinal vascular endothelium. Invest Ophthalmol Vis Sci. 2013;54:6917–6925. doi: 10.1167/iovs.13-12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith JR, Choi D, Chipps TJ, Pan Y, Zamora DO, Davies MH, et al. Unique gene expression profiles of donor-matched human retinal and choroidal vascular endothelial cells. Invest Ophthalmol Vis Sci. 2007;48:2676–2684. doi: 10.1167/iovs.06-0598. [DOI] [PubMed] [Google Scholar]
- 8.Ashander LM, Appukuttan B, Ma Y, Gardner-Stephen D, Smith JR. Targeting endothelial adhesion molecule transcription for treatment of inflammatory disease: a proof-of-concept study. Mediators Inflamm. 2016;2016:7945848. doi: 10.1155/2016/7945848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo W, Liu S, Cheng Y, Lu L, Shi J, Xu G, et al. ICAM-1-Related noncoding RNA in cancer stem cells maintains ICAM-1 expression in hepatocellular carcinoma. Clin Cancer Res. 2016;22:2041–2050. doi: 10.1158/1078-0432.CCR-14-3106. [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Hu HS, Feng YX, Shi J, Li N, Guo WX, et al. Characterisation of a novel cell line (CSQT-2) with high metastatic activity derived from portal vein tumour thrombus of hepatocellular carcinoma. Br J Cancer. 2010;102:1618–1626. doi: 10.1038/sj.bjc.6605689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulyk ML. Computational prediction of transcription-factor binding site locations. Genome Biol. 2003;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, et al. Ensembl 2018. Nucleic Acids Res. 2017;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 15.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K, Brown EE, Choi CB, Alarcon-Riquelme ME, BIOLUPUS. Kelly JA, et al. Variation in the ICAM1-ICAM4-ICAM5 locus is associated with systemic lupus erythematosus susceptibility in multiple ancestries. Ann Rheum Dis. 2012;71:1809–1814. doi: 10.1136/annrheumdis-2011-201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egerer K, Feist E, Rohr U, Pruss A, Burmester GR, Dorner T. Increased serum soluble CD14, ICAM-1 and E-selectin correlate with disease activity and prognosis in systemic lupus erythematosus. Lupus. 2000;9:614–621. doi: 10.1191/096120300678828749. [DOI] [PubMed] [Google Scholar]
- 19.Kammerer S, Roth RB, Reneland R, Marnellos G, Hoyal CR, Markward NJ, et al. Large-scale association study identifies ICAM gene region as breast and prostate cancer susceptibility locus. Cancer Res. 2004;64:8906–8910. doi: 10.1158/0008-5472.CAN-04-1788. [DOI] [PubMed] [Google Scholar]
- 20.Pare G, Chasman DI, Kellogg M, Zee RY, Rifai N, Badola S, et al. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 2008;4:e1000118. doi: 10.1371/journal.pgen.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pare G, Ridker PM, Rose L, Barbalic M, Dupuis J, Dehghan A, et al. Genome-wide association analysis of soluble ICAM-1 concentration reveals novel associations at the NFKBIK, PNPLA3, RELA, and SH2B3 loci. PLoS Genet. 2011;7:e1001374. doi: 10.1371/journal.pgen.1001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu ZG. Molecular mechanism of TNF signaling and beyond. Cell Res. 2005;15:24–27. doi: 10.1038/sj.cr.7290259. [DOI] [PubMed] [Google Scholar]
- 23.Brenner D, Blaser H, Mak TW. Regulation of tumour necrosis factor signalling: live or let die. Nat Rev Immunol. 2015;15:362–374. doi: 10.1038/nri3834. [DOI] [PubMed] [Google Scholar]
- 24.Appukuttan B, Ashander LM, Ma Y, Smith JR. Selection of reference genes for studies of human retinal endothelial cell gene expression by reverse transcription-quantitative real-time polymerase chain reaction. Gene Rep. 2018;10:123–134. doi: 10.1016/j.genrep.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(− delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Sources for eutherian genomes used in genome evolutionary rate profiling (GERP) and chromosomal locations of GERP-constrained elements.
Additional file 2: Table S2. Predicted transcription factor binding sites within genome evolutionary rate profiling (GERP)-constrained elements.
Additional file 3: Table S3. Description of phenotype-nucleotide polymorphisms (SNPs) predicted to influence transcription of the human ICR gene.
Additional file 4: Table S4. Primer pairs and product sizes for gene transcripts studied in human retinal endothelial cells. References are provided for primers sequences sourced from the literature.
Data Availability Statement
All data generated or analyzed during this study are included in the manuscript.


