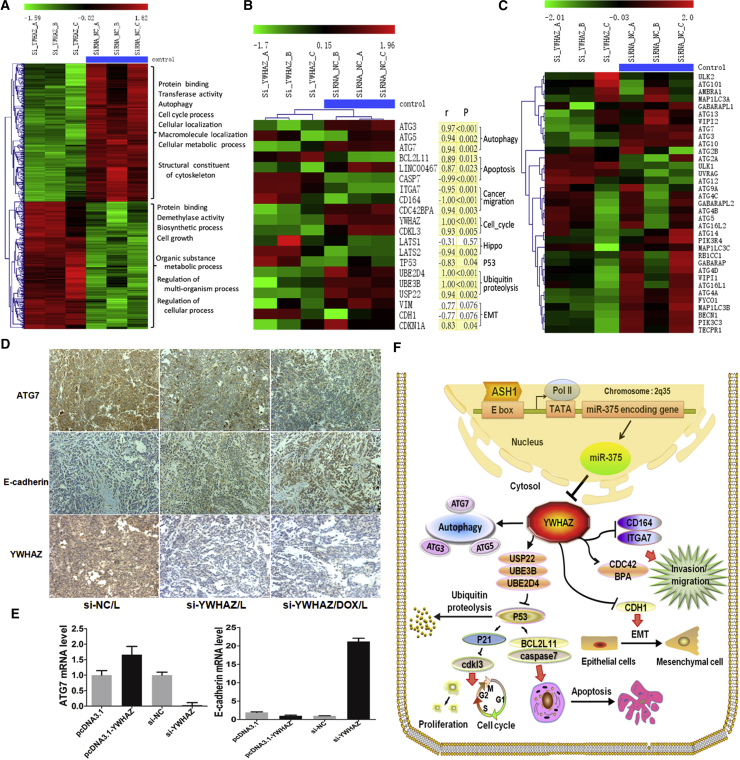
Figure 7.
Potential Mechanism and Downstream Pathways of the ASH1-miR-375-YWHAZ Axis in HCC
(A) Heatmap of 645 differentially expressed genes from si-YWHAZ microarray. Each row of gene probes was normalized, and each column sample was clustered by hierarchical clustering. Two different state samples were found to be clustered together (the first three columns were YWHAZ knockout samples, and the last three were control samples [blue scale]). The uppermost scale from green to red indicates that the expression level is low to high. The main function and biological processes of the up and down genes are also shown in the figure. (B) The expression profiles of YWHAZ downstream genes from si-YWHAZ microarray are shown. The heatmap genes were sorted according to different pathways, and most of these genes were significantly differently expressed between si-YWHAZ samples and control. p, significance measure of the correlation; r, Pearson correlation coefficient of each gene and YWHAZ expression. Shading yellow represents the result is significant; otherwise, no color. The results showed that most of these genes were significantly correlated with YWHAZ expression (p < 0.05). (C) Heatmap of 35 autophagy genes in si-YWHAZ microarray data is shown. (D) Immunohistochemistry of ATG7, E-cadherin, and YWHAZ in the three groups of mice tumor tissues treated with siRNA-NC/L, si-YWHAZ/L, or si-YWHAZ/DOX/L is shown. (E) The expressions of ATG7 and E-cadherin were detected in SMMC-7721 cells after transfection by SYBR green qRT-PCR. (F) Schematic illustration of ASH1-miR-375-YWHAZ signaling axis and its possible downstream pathways in hepatocellular carcinoma is shown.