Abstract
β-Lactam antibiotics are widely used to treat urinary tract infections in Nigeria. This study aimed to determine the presence and characteristics of extended spectrum β-lactamases in commonly isolated uropathogenic Gram-negative bacteria (GNB) in Nigeria.
Fifty non-duplicate GNB isolates consisting of Escherichia coli, 19; Klebsiella pneumoniae, 21; and Pseudomonas aeruginosa, 10 were obtained from three tertiary hospitals in Nigeria. The antibiotic susceptibility testing of all isolates to a panel of antibiotics including minimum inhibitory concentrations (MICs) and extended spectrum β-lactamases was determined. Polymerase chain reactions and sequencing were used to detect β-lactam genes.
Polymerase chain reactions and sequencing identified varying extended spectrum β-lactamases (ESBLs) encoding genes for 24 isolates (48.0%). Cefotaximase-Munich (CTX-M) 15 was the dominant gene with 20/24 of the isolates positive at 83.3%; multiple genes (2 to 6 ESBL genes) were found in 20 of the isolates. The isolates encoded other genes such as CTX-M-14, 33.3%; sulfhydryl variable (SHV) variants, 58.3%; oxacillinase (OXA) variants, 70.8%; OXA-10, 29.2%; and Vietnamese extended β-lactamase (VEB) 1, 25.0%. There was no difference between the MIC50 and MIC90 of all the isolates.
The high-level multidrug resistance of uropathogens to third generation cephalosporins including other antibiotics used in this study is strongly associated with carriage of ESBLs, predominantly CTX-M-15, as well as CTX-X-M-14, OXA-10, and VEB-1.
Keywords: extended spectrum β-lactamases, uropathogens, antibiotics, resistance, Nigeria
Introduction
Extended spectrum β-lactamases (ESBLs) are enzymes produced by a variety of Gram-negative bacteria which confer an increased resistance to commonly used antibiotics, such as the penicillins and the cephalosporins [1]. These ESBLs are mostly plasmid mediated and efficiently hydrolyze oxyimino cephalosporins (ceftazidime, cefotaxime, and ceftriaxone) and monobactams. They are inhibited by β-lactamase inhibitors such as clavulanate, sulbactam, and tazobactam [2]. Plasmids coding for ESBLs may also carry additional β-lactamase genes as well as genes conferring resistance to other antimicrobial classes [3, 4]. This can limit the chemotherapeutic options for ESBL-producing pathogens and facilitates inter-species and intra-species dissemination of ESBLs [5]. As such, they are a worrying global public health issue as infections caused by such enzyme-producing organisms are associated with a higher morbidity and mortality with greater fiscal burden coupled with increasing prevalence rates worldwide and an ever diminishing supply in the antibiotic armamentarium, and these enzymes represent a clear and present danger to public health [1].
ESBLs have been found mainly in Enterobacteriaceae, particularly Klebsiella species and Escherichia coli, but have also been reported in other genera, such as Citrobacter, Enterobacter, Morganella, Proteus, Providencia, Salmonella, and Serratia and in Pseudomonaceae. In Nigeria urinary tract infection (UTI) is one of the most common diseases that make people seek medical attention with the majority of infections caused by Enterobacteriaceae and Pseudomonaceae [6]. Reports of multidrug-resistant isolates have increased during the last decade, probably as a result of the extensive use of broad-spectrum antibiotics. Cefotaximase-Munich (CTX-M)-type ESBLs have emerged among E. coli and Klebsiella pneumoniae and are now commonly isolated from UTIs, and often, these enzymes are carried by strains with a multidrug resistance phenotype. Multidrug resistance expressed by CTX-M-producing isolates from the community is often associated with the presence of multiple ESBLs genes, as well as aminoglycoside and quinolone resistance genes, thereby limiting the choice of effective antimicrobial drugs. Studies on the causative agents of UTIs in Nigeria and their susceptibility to antimicrobials are lacking; particularly, there is paucity of information on the characterization of ESBLs genes in Enterobacteriaceae and Pseudomonaceae from patients with UTIs. Therefore, the study aimed to determine the presence and characteristics of extended spectrum β-lactamases in commonly isolated uropathogenic GNB in Nigeria.
Materials and Methods
Bacterial Isolates and Study Sites. A total of 50 non-duplicate unbiased GNB isolates consisting of E. coli, 19; K. pneumoniae, 21; and Pseudomonas aeruginosa, 10 were obtained from three tertiary hospitals in Nigeria. The distribution of the isolates from the hospitals is as follows: hospital I, 24; hospital II, 16; and hospital III, 10; these hospitals are located at Ibadan, Ogbomoso, and Osogbo, respectively, in South West Nigeria. Each isolate was obtained from a patient with significant bacteriuria diagnosed of varying specific and general UTIs ranging from pyelonephritis, cystitis, prostatitis, and so on. All isolates were speciated using Analytical Profile Index (API) 20E strips (BioMerieux, France) and conventional biochemical tests. The study was carried out at the Molecular Biology Laboratory of Department of Biomedical Science, Ladoke Akintola University of Technology, Ogbomoso, Osogbo campus.
Antibiotic Susceptibility Testing. The antibiotic susceptibility patterns of all the isolates to a panel of antibiotics were determined by the disk diffusion method (disks from Oxoid, UK) using Mueller-Hinton agar according to Clinical Laboratory Standards Institute (CLSI) guidelines [7]. Resistant isolates were further selected for susceptibility testing to β-lactam drugs such as ceftriaxone, ceftazidime or Amoxycillin-clavulanic acid using the agar doubling dilution method [8]. All runs included the control organisms such as E. coli (NCTC 10418) for Enterobacteriaceae and P. aeruginosa (NCTC 10662) for Pseudomonas species.
Phenotypic Detection of β-lactamases. Broth cultures of each test strain as well as ampicillin-resistant E. coli strain NCTC 10418 (carrying pUC18) were incubated overnight in 5 ml of Luria–Bertani broth. Then, 200 μl of the resulting overnight culture was transferred into the wells of a microtiter tray, and sterile broth was included as a negative control. Then, 10 μl of nitrocefin solution (Fisher Scientific, Loughborough, UK) prepared according to the manufacturer's instructions was added to each well. β-Lactamase production was inferred when the broth turned red within 30 min of addition of nitrocefin as directed by the manufacturer.
Detection of Extended-spectrum β-lactamases by the Double-disk Diffusion Test (DDDT). Suspensions of each test strain were prepared in sterile water to give an inoculum equivalent to a 0.5 McFarland standard before being used to inoculate the surface of Mueller-Hinton agar plates [8]. ESBL-positive K. pneumoniae ATCC 700603 and ESBL negative E. coli ATCC 25922 control strains were used in these experiments. A 30-μg ceftazidime disk (the best indicator for Temoneira (TEM)- and SHV-derived ESBLs) was placed on the left of each plate and a 30-μg cefotaxime disk (the best indicator for CTX-M types) was placed on the right; an AMC (20/10 μg) (Oxoid Ltd., Basingstoke, UK) was placed in the center between the other disks. The disks were placed 25–30 mm apart, center-to-center. Following overnight incubation in air at 37 °C, ESBL production was inferred when the zone of inhibition around the ceftazidime and cefotaxime disks was expanded by ≥5 mm by the presence of clavulanic acid.
Amplification of ESBL Genes. Polymerase chain reaction (PCR) was used to detect genes encoding resistance to β-lactams (blaOXA, blaSHV, blaCTX-M, blaVEB, blaPER, blaOXA-10, blaOXA-48, blaKPC, blaNDM, blaGES, blaVIM) as previously described (Table 1) [9, 10]. Amplimers resulting from these PCR reactions were sequenced at Institute of Microbiology and Infection, University of Birmingham, Birmingham, UK to confirm the identity and specific variant of each gene identified and sequences were aligned to known reference sequences using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/help/index.html).
Table 1.
Primer used for the amplification of β-lactamase genes
| Forward primer | Sequence (5′-3′) | Reverse primer | Sequence (3′-5′) | Annealing temp. (°C) | Product size (bp) |
|---|---|---|---|---|---|
| CTX-M-1 | GACGATGTCACTGGCTGAGC | CTX-M-1 | AGCCGCCGACGCTAATACA | 60 | 499 |
| CTX-M-9 | GCTGGAGAAAAGCAGCGGAG | CTX-M-9 | GTAAGCTGACGCAACGTCTG | 60 | 293 |
| SHV | AGGATTGACTGCCTTTTTG | SHV | ATTTGCTGATTTCGCTCG | 56 | 393 |
| OXA | ATATCTCTACTGTTGCATCTCC | OXA | AAACCCTTCAAACCATCC | 50 | 216 |
| OXA-10 | GTCTTTCGAGTACGGCATTA | OXA-10 | ATTTTCTTAGCGGCAACTTAC | 52 | 600 |
| OXA-48 | TTCGGCCACGGAGCAAATCAG | OXA-48 | GATGTGGGCATATCCATATTCATCGCA | 56 | 240 |
| PER-1 | ATGAATGTCATTATAAAAGC | PER-1 | AATTTGGGCTTAGGGCAGAA | 51 | 590 |
| VEB | CGACTTCCATTTCCCGATGC | VEB | GGACTCTGCACCAAATACGC | 55 | 604 |
| KPC | ATGTCACTGTATCGCCGTCT | KPC | TAGACGGCCAACACAATAGG | 56 | 785 |
| NDM | TTGATGCTGAGCGGGTG | NDM | CTGTCCTTGATCAGGCAGC | 56 | 578 |
| VIM | AGTGGTGAGTATCCGACAG | VIM | ATGAAAGTGCGTGGAGAC | 56 | 261 |
| GES | CGGTTTCTAGCATCGGGACACAT | GES | CCGCCATAGAGGACTTTAGCACAG | 58 | 263 |
RAPD PCR Typing. The epidemiological relationships between multiple strains of E. coli, K. pneumoniae, and P. aeruginosa carrying ESBLs were analyzed according to species by randomly amplified polymorphic DNA (RAPD). The primers sequence and PCR running conditions used were according to Vogel et al. [11] modified to use 1 μl of 100 μmol of primers at a final concentration of 0.02 μmol [6]. The experiment was repeated twice for each species to ensure reproducibility. DNA fingerprints were compared by visual inspection to assign similar banding patterns to the same RAPD type.
Statistics Analysis. Data were analyzed using the statistical package within Microsoft Excel. χ2 was used to determine the association between distribution of ESBL genes and hospitals. In this case, p value less than 0.05 was considered to be significant.
Results
Antimicrobial Susceptibility Testing. A total of 50 clinical isolates of GNB were obtained from three tertiary hospitals in South West Nigeria. These isolates were obtained from patients suffering from UTIs with significant bacteriuria. API and other biochemical tests characterized these GNB to be E. coli, K. pneumoniae, and P. aeruginosa. The results of antimicrobial susceptibility testing showed that colistin was more susceptible at 78% followed by imipenem, 72%. The cephalosporin class of antimicrobial agent such as ceftriaxone recorded 32% with ceftazidime having 30%. Surprisingly, tigecycline had the least susceptibility of 0% followed by ciprofloxacin 2% (Table 2). The minimum inhibitory concentrations (MICs) of ceftriaxone, ceftazidime, and amoxycillin-clavulanic acid showed that there were no differences between the MIC50 and MIC90 of all the isolates. MIC50 and MIC90 of K. pneumoniae, E. coli, and P. aeruginosa were >128 μg/ml having 100% resistance in some cases (Table 3). These results indicate that the uropathogenic isolates were highly resistant strains.
Table 2.
Summary of antimicrobial disk susceptibility testing of 50 bacterial isolates
| Antibiotics (μg/ml) | Sensitive | Intermediate | Resistance |
|---|---|---|---|
| Imipenem (10) | 36(72) | 4(8) | 10(20) |
| Meropenem (10) | 21(42) | 12(24) | 17(34) |
| Amoxycillin-clavulanic | 9(18) | 5(10) | 37(74) |
| acid (30) | |||
| Ceftazidime (30) | 15(30) | 5(10) | 30(60) |
| Cefotaxime (30) | 10(20) | 3(6) | 37(74) |
| Cefpodoxime (10) | 11(22) | 2(4) | 37(74) |
| Ceftriaxone (30) | 16(32) | 2(4) | 32(64) |
| Nalidixic acid (30) | 8(16) | 2(4) | 40(80) |
| Ciprofloxacin (5) | 1(2) | 7(14) | 42(84) |
| Chloramphenicol | 19(38) | 1(2) | 30(60) |
| Colistin | 38(78) | 0 (0) | 11(22) |
| Sulfonamide | 6(12) | 1(2) | 43(86) |
| Tigecycline | 0 (0) | 3(6) | 47(94) |
Numbers in parentheses are percentages
Table 3.
Minimum inhibitory concentrations (MICs) of the 50 bacterial isolates
| Isolate, n | Antimicrobial agents | MIC (0.06–128 μg/ml) | Sensitive | Intermediate | Resistant | |
|---|---|---|---|---|---|---|
| MIC50 | MIC90 | |||||
| Escherichia coli, 19 | Ceftriaxone | >128 | >128 | 0 (0.00) | 1 (5.2) | 18 (94.7) |
| Ceftazidime | >128 | >128 | 0 (0.00) | 3 (15.7) | 15 (78.9) | |
| Amoxycillin-clavulanic acid | >128 | >128 | 0 (0.00) | 0 (0.00) | 19(100) | |
| Klebsiella pneumoniae, 21 | Ceftriaxone | >128 | >128 | 1 (4.8) | 0 (0.00) | 20 (95.0) |
| Ceftazidime | >128 | >128 | 0 (0.00) | 0 (0.00) | 21(100) | |
| Amoxycillin-clavulanic acid | >128 | >128 | 0 (0.00) | 0 (0.00) | 21(100) | |
| Pseudomonas aeruginosa, 10 | Ceftriaxone | >128 | >128 | 0 (0.00) | 0 (0.00) | 10(100) |
| Ceftazidime | >128 | >128 | 0 (0.00) | 0 (0.00) | 10(100) | |
| Amoxycillin-clavulanic acid | >128 | >128 | 0 (0.00) | 0 (0.00) | 10(100) | |
Numbers in parentheses are percentages
Detection of Extended Spectrum β-lactamases. Twenty-one (42%) of the isolates were ESBL producers (Table 4). Polymerase chain reactions and sequencing identified varying ESBLs encoding genes for 24 isolates (48.0%). CTX-M-15 was the dominant gene with 20/24 of the isolates positive at 83.3%; multiple genes (2 to 6 ESBL genes) were found in 20 of the isolates. The isolates encoded other genes such as CTX-M-14, 33.3%; SHV variants, 58.3% (all SHV variants were encoded as a multiple gene); oxacillinase (OXA) variants, 70.8%; OXA-10, 29.2%; and Vietnamese extended β-lactamase (VEB) 1, 25.0%. Of these, 11/19 (57.9%) E. coli, 11/21 (52.4%) K. pneumoniae, and 2/10 (20%) P. aeruginosa carried these respective ESBL genes. However, none of the isolates was positive for any of the carbapenemase genes tested in this study. The ESBL genes were found in the three hospitals and the isolates in varying proportions including the clinical diagnosis. However, no association was found between hospitals and distribution of ESBL genes (X2 = 1.32; p = 0.85; p > 0.05).
Table 4.
Carriage of β-lactamase genes and their minimum inhibitory concentrations
| ID no. | Clinical diagnosis | Isolate | MIC (μg/ml) | ESBL phenotype (DDST) | ESBLs genes | ||
|---|---|---|---|---|---|---|---|
| CAZ | CRO | AMC | |||||
| AR7 | UTI in pregnancy | E. coli | >128 | >128 | >128 | - | CTX-M-15, OXA |
| AR8 | UTI | E. coli | >128 | >128 | >128 | - | OXA |
| AR11 | Pelvic inflammatory disease | E. coli | 128 | >128 | >128 | - | OXA |
| AR12 | Prostate enlargement | E. coli | >128 | >128 | >128 | - | CTX-M-15, CTX-M-14 |
| AR20 | UTI | E. coli | >128 | >128 | >128 | + | CTX-M-15, CTX-M-14, SHV, OXA, OXA-10, VEB-1 |
| AR28 | Cystitis | E. coli | >128 | >128 | >128 | + | CTX-M-15, SHV, VEB-1, OXA |
| AR31 | Prostatitis | E. coli | 16 | 128 | >128 | + | CTX-M-15, CTX-M-9, SHV, OXA and OXA-10 |
| AR33 | Pelvic inflammatory disease | E. coli | 8 | 8 | >128 | - | OXA, SHV and OXA-10 |
| AR51 | UTI | E. coli | >128 | >128 | >128 | + | CTX-M-15 |
| AR65 | UTI | E. coli | >128 | >128 | >128 | + | CTX-M-15, CTX-M-14, OXA and OXA-10 |
| AR73 | Urosepsis | E. coli | 64 | >128 | >128 | + | CTX-M-15, SHV |
| AR2 | Prostatitis | K. pneumoniae | 32 | >128 | >128 | - | CTX-M-15, SHV, OXA |
| AR16 | Chronic kidney disease | K. pneumoniae | >128 | >128 | >128 | - | CTX-M-15, SHV, OXA |
| AR17 | Burn injury | K. pneumoniae | >128 | >128 | >128 | - | SHV, OXA |
| AR18 | UTI | K. pneumoniae | >128 | >128 | >128 | + | CTX-M-15, CTX-M-14, SHV, OXA, OXA-10, VEB-1 |
| AR23 | UTI | K. pneumoniae | >128 | >128 | >128 | - | CTX-M-15, SHV, OXA |
| AR32 | Glomerulonephritis | K. pneumoniae | 128 | >128 | >128 | - | CTX-M-15 |
| AR54 | Prostate enlargement | K. pneumoniae | >128 | >128 | >128 | + | CTX-M-15, CTX-M-14, SHV, OXA |
| AR55 | Urosepsis | K. pneumoniae | >128 | >128 | >128 | + | CTX-M-15, CTX-M-14, SHV, OXA, OXA-10, VEB-1 |
| AR71 | Prostatitis | K. pneumoniae | >128 | >128 | >128 | - | CTX-M-15, CTX-M-14 |
| AR78 | Pelvic inflammatory disease | K. pneumoniae | 128 | >128 | >128 | + | CTX-M-15, CTX-M-14 |
| AR79 | UTI | K. pneumoniae | 32 | >128 | >128 | + | CTX-M-15, SHV- |
| AR26 | Pyelonephritis | P. aeruginosa | >128 | >128 | >128 | + | CTX-M-15, SHV, OXA and VEB-1 |
| AR68 | Pyelonephritis | P. aeruginosa | >128 | >128 | >128 | + | CTX-M-15, OXA, OXA-10, VEB-1 |
CTX-M-1, cefotaximase-Munich-1; OXA, oxacillinase; VEB, Vietnamese extended β-lactamase; MIC, minimum inhibitory concentration; EC, Escherichia coli; KP, Klebsiella pneumoniae; PA, Pseudomonas aeruginosa; DDST, double disc synergyst; +, positive; –, negative
Typing of Isolates. The degree of clonality among the resistant isolates revealed high diversity among all the isolates tested with no identical RAPD or banding patterns observed except lanes 3 and 5 (Figure 1). This suggests that the resistance is underpinned by spread of resistance genes and not expansion of a dominant clone(s).
Figure 1.
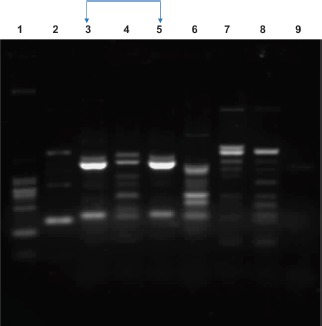
Representative RAPD typing of E. coli isolates showing different banding patterns. Lanes 1 to 9 showed the banding patterns of each isolates; no identical banding patterns were observed except for lanes 3 and 5
Discussion
CTX-M-15 was the dominant ESBL gene, followed by OXA, SHV, CTX-M-14, OXA-10, and VEB-1 in frequent order. This is consistent with the present antimicrobial resistance situation among Enterobacteriaceae where the CTX-M family has replaced TEM and SHV types and became the dominant ESBL in most parts of the world including Nigeria, where they are prevalent both in the hospital and community settings [6, 12, 13]. In Morocco, CTX-M was detected in 6 out of 7 ESBL-producing E. coli with a predominance of CTX-M-15 (6/6) [14], while in Cameroon all the ESBL-E. coli strains isolated from stool samples of women with UTIs contained group 1 CTX-M enzymes [15]. Similarly, CTX-M-15 dominated ESBLs in Enterobacteriaceae isolated from environmental samples in a hospital in Tunisia [16]. The high distribution of CTX-M-15 type ESBLs among these isolates explains the high rate of resistance to cephalosporin such as cefotaxime, ceftriaxone, and ceftazidime in our study.
CTX-M-15 co-existed with either OXA-10 or VEB-1 in all the isolates of E. coli, Klebsiella species, and P. aeruginosa. VEB-1 was first described in an E. coli isolate from Vietnam in 1998 [17]. Ever since, it has been reported from various parts of the world in different species of Enterobacteriaceae [18], including Nigeria where Aibinu et al. reported the emergence of OXA-10, VEB-1, and Cephamycins (CMY) β-lactamases and mobile elements of clinical Providencia isolates from a catheter tip of a patient in Lagos [19]. To the best of our knowledge, this is the first report of VEB-1 and OXA-10 in clinical isolates of E. coli, Klebsiella species, and P. aeruginosa from Nigeria. OXA-10 is an important serine β-lactamase because it is inhibited only weakly by clavulanic acid. Interestingly, the risk of widespread ESBL-producing strains among uropathogens not only rendered oxyimino-cephalosporins ineffective but also pose a therapeutic challenge since they are frequently resistant to other kinds of antimicrobial drugs, including aminoglycosides, quinolones, and co-trimoxazole [20]. There is no association between ESBL genes in the isolates and the various hospitals suggesting that the resistance genes spread across without any peculiar determinant and the RAPD typing found no major clonal or epidemiological relationship which suggested the occurrence and dissemination of a plasmid blaESBL.
It is common knowledge that production of ESBLs is the most common mechanism of resistance to third-generation cephalosporins among Enterobacteriaceae including K. pneumoniae and E. coli and mediates multidrug resistance as shown in this study. There is high-level resistance to various other antibiotics including the unprecedented high-level resistance to tigecycline and the level of resistance to colistin and meropenem, a last resort carbapenem antibiotic against GNB. These findings have significant implications for the management of patients with urinary tract infections using third-generation cephalosporins or fluroquinolones, including carbapenems. Carbapenem resistance development in isolates producing CTX-M and other ESBLs due to selection of mutants lacking expression of outer membrane porins has been reported [21, 22]. Resistance of UTI pathogens to the panels of antibiotics may not be unconnected with their frequent prescription in the hospital, their easy availability in the community without prescription, and their relative affordability which make them subject to abuse.
Conclusions
The high-level multidrug resistance of uropathogens to third-generation cephalosporins including other antibiotics used in this study is strongly associated with carriage of ESBLs found in this study, predominantly CTX-M-15, as well as CTX-M-14, OXA-10, and VEB-1. A combination therapy is recommended for use guided by antimicrobial susceptibility testing, and we should furthermore exercise restraint in using or prescribing carbapenems or colistin to prevent selection of resistant isolates.
Acknowledgments
The authors would like to thank the Medical Laboratory Scientists from the hospitals where assembly of bacterial isolates were collected for their cooperation and the technical staff in our department.
Footnotes
Funding sources
No financial report was received for this study.
Authors' contribution
D.O.O. has the study concept, designed the work, interpreted the data, and prepared the article. O.A.T.A. codesigned, supervised, and prepared the article. M.A.W. codesigned and analyzed certain aspect of the data. A.S.O. supervised and also contributed in data analysis. O.M.O. analyzed the data and prepared the article. All authors have full access to all data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Dhillon RHP, Clark J. ESBL: A clear and present danger? Crit Care Res Pract. 2012;2012:625170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush K, Fisher JF. Epidemiological expansion, structural studies and clinical challenges of new beta-lactamases from gram-negative bacteria. Ann Rev Microbiol. 2011;65:455–78. [DOI] [PubMed] [Google Scholar]
- 3.Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8:159–66. [DOI] [PubMed] [Google Scholar]
- 4.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zahar JR, Lortholary O, Martin C, Potel G, Plesiat P, Nordmann P. Addressing the challenge of extended-spectrum beta-lactamases. Curr Opin Investig Drugs. 2009;10:172–80. [PubMed] [Google Scholar]
- 6.Ogbolu DO, Webber MA. High-level and novel mechanisms of carbapenem resistance in Gram-negative bacteria from tertiary hospitals in Nigeria. Int J Antimicrob Agents. 2014;43:412–7. [DOI] [PubMed] [Google Scholar]
- 7.Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty Fifth Informational Supplement, M100-S25. Wayne, PA, USA: CLSI; 2015. [Google Scholar]
- 8.Andrew JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48;5–16. [DOI] [PubMed] [Google Scholar]
- 9.Maynard C, Fairbrother JM, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, et al. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob Agents Chemother. 2003;47:3214–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA. Hooper DC: qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother. 2006;50:2872–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel L, Jories G, Tviep S, Koek A, Dijkshoorn L. RAPD typing of Klebsiella pneumoniae, Klebsiella oxytoca, Serratia marcescens and Pseudomonas aeruginosa isolates using standardized reagents. Clin Microbiol Infect. 1999; 5:270–6. [DOI] [PubMed] [Google Scholar]
- 12.Livermore DM. Current epidemiology and growing resistance of Gram-negative pathogens. Korean J Intern Med. 2012;27:128–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogbolu DO, Webber MA. High level and novel mechanisms of carbapenem resistance in Gram negative bacteria from tertiary hospitals in Nigeria. Int J Antimicrob Agents. 2014;43:412–7. [DOI] [PubMed] [Google Scholar]
- 14.Bourjilat F, Bouchrif B, Dersi N, Claude JDPG, Amarouch H, Timinouni M. Emergence of extended-spectrum beta-lactamase-producing Escherichia coli in community-acquired urinary infections in Casablanca, Morocco. J Infect Dev Ctries. 2011;5:850–5. [DOI] [PubMed] [Google Scholar]
- 15.Djuikoue I, Woerther P, Toukam M, Burdet C, Ruppé E, Gonsu K, et al. Intestinal carriage of extended spectrum beta-lactamase producing E. coli in women with urinary tract infections, Cameroon. J Infect Dev Ctries. 2016;10:1135–9. [DOI] [PubMed] [Google Scholar]
- 16.Dziri R, Klibi N, Alonso CA, Said LB, Bellaaj R, Slama KB, et al. Characterization of extended-spectrum β-lactamase (ESBL)-producing Klebsiella, Enterobacter, and Citrobacter obtained in environmental samples of a Tunisian hospital. Diagn Infect Microbiol Dis. 2016;86:190–3. [DOI] [PubMed] [Google Scholar]
- 17.Naas T, Benaoudia F, Massuard S, Nordmann P. Integron-located VEB-1 extended-spectrum beta-lactamase gene in a Proteus mirabilis clinical isolate from Vietnam. J Antimicrob Chemother. 2000;46:703–11. [DOI] [PubMed] [Google Scholar]
- 18.Jain S, Gaind R, Kothari C, Sehgal R, Shamweel A, Thukral SS, et al. VEB-1 extended-spectrum β-lactamase producing multidrug-resistant Proteus mirabilis sepsis outbreak in a neonatal intensive care unit in India: clinical and diagnostic implications. JMM Case Rep. 2016;3:e005056 DOI: 10.1099/jmmcr 0.005056 eCollection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aibinu I, Pfeifer Y, Ogunsola F, Odugbemi T, Koenig W, Ghebremedhin B, et al. Emergence of beta-lactamases OXA-10, VEB-1 and CMY in Providencia spp. from Nigeria. J Antimicrob Chemother. 2011;66:1931–2. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Liu WE, Li H, Duan H, Zhang Y, Liang X, et al. Novel CTX-M β-lactamase genotype distribution and spread into multiple species of Enterobacteriaceae in Changsha, Southern China. J Antimicrob Chemother. 2009;68:245–8. [DOI] [PubMed] [Google Scholar]
- 21.Cornaglia G, Russell K, Satta G, Fontana R. Relative importances of outer membrane permeability and group 1 beta-lactamase as determinants of meropenem and imipenem activities against Enterobacter cloacae. Antimicrob Agents Chemother. 2012;39:350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Fernandez A, Miriagou V, Papagiannitsis CC, Giordano A, Venditti M, Mancini C, Carattoli A. An ertapenem-resistant extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob Agents Chemother 2010;54:4178–84. [DOI] [PMC free article] [PubMed] [Google Scholar]


