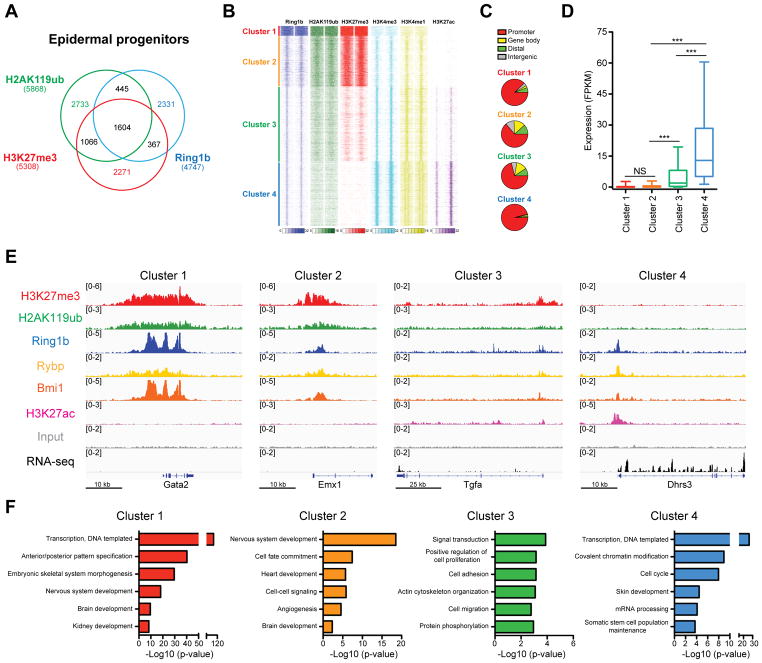
Figure 1. Genome-wide analysis of PRC1 binding reveals enrichment at both active and repressed genes.
(A) Overlap between Ring1b, H2AK119ub, and H3K27me3 ChIP-Seq peaks in epidermal progenitors. (B) k-means clustering of ChIP-seq read densities across Ring1b-peaks only, for duplicates of Ring1b, and indicated histone modifications, with input signal subtracted. (C) Genomic distribution of Ring1b peaks (D) Expression analysis of genes located near Ring1b peaks in control epidermal progenitors. n=3. ***p<0.001; NS, not significant (one-way ANOVA with a Bonferroni post-hoc test). (E) IGV browser views of Ring1b, H2AK119ub, H3K27me3, H3K27ac, Rybp, Bmi1, and input for indicated representative genes for each Ring1b-bound cluster. (F) Gene ontology (GO) analysis for Ring1b target genes. See also Figure S1 and Table S1.