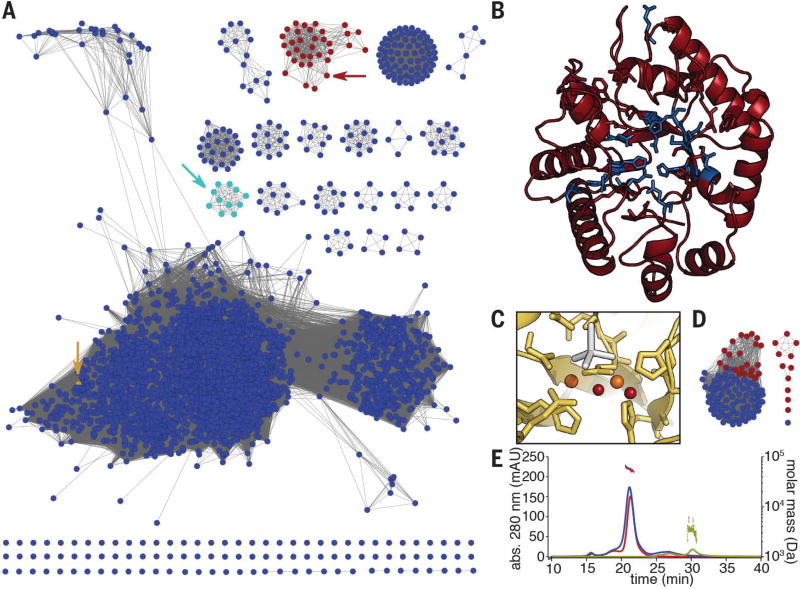
Fig. 2. Bioinformatic and biochemical analyses of the MbnBC heterodimer.
(A) Sequence similarity network for the DUF692 family. MbnBs are depicted in red, MbnXs (found only in the fifth Mbn subgroup) are in teal, and the structurally characterized Hs. somnus 129Pt protein sequence (PDB: 3BWW) is in yellow. Arrows indicate the location of the relevant nodes. (B) The predicted structure of Ms. trichosporium OB3b MbnB (red), as modeled against 3BWW using iTasser (RMSD, 4.2 ± 2.8 Å). Residues in blue are strictly conserved in MbnBs. (C) Diiron site as observed in the 3BWW structure (yellow). All amino acids coordinating the two irons (orange) are strictly conserved in MbnBs. Two water molecules (red) are also present in the coordination sphere in the crystal structure, along with a cacodylate molecule (gray) from the crystallization buffer. (D) Sequence similarity network for the broader MbnC family. Red circles, MbnCs; blue circles, related non-MbnC genes found exclusively in Pseudomonas species. (E) SEC-MALS analysis of MbnA (yellow; 3 kDa predicted, 3 kDa observed), MbnBC (red; 58 kDa predicted, 55 kDa observed), and MbnABC (blue; 61 kDa predicted, 58 kDa observed) indicates that MbnB and MbnC form a heterodimeric complex that can bind MbnA.