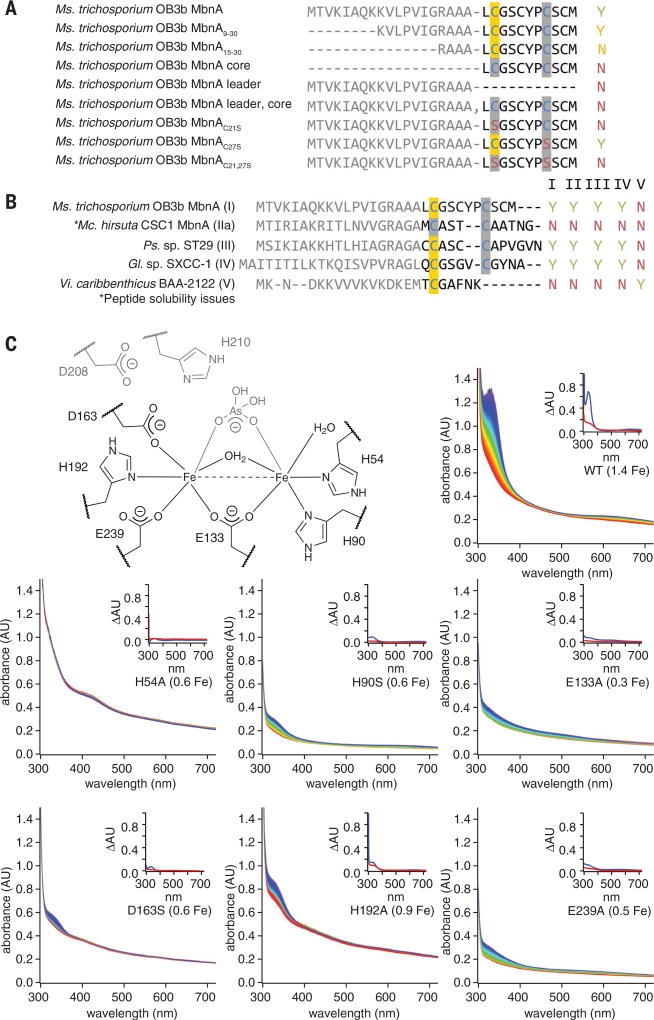
Fig. 4. Correlation of MbnBC activity with proposed diiron active site ligands and MbnA sequence.
(A) Summary of activity observed via UV-vis absorption spectroscopy and LC-MS for 125 µM Ms. trichosporium OB3b MbnA variants with 100 µM Ms. trichosporium OB3b MbnBC and excess O2. Yes (Y) indicates that modification is observed; No (N) indicates no modification. Modified cysteines localized via tandem MS are highlighted in yellow. (B) Summary of activity observed via UV-vis absorption spectroscopy and LC-MS for 125 µM MbnA substrate peptides and 100 µM MbnBC enzyme complexes from the different Mbn operon families in the presence of excess (~900 µM) O2. (C) Stopped-flow UV-vis absorption spectra over a period of 250 s for reactions between 125 µM MbnA, excess (~900 µM) O2, and 100 µM wild-type Ms. trichosporium OB3b MbnBC or one of six enzyme variants with alanine mutations in predicted active-site residues (the diiron site from the Hs. somnus 129Pt structure, labeled with Ms. trichosporium OB3b residue numbers, is provided for reference). Spectra are colored in rainbow order according to time, with the earliest spectra in red and the latest spectra in blue. Iron content per heterodimer is included in each label. Insets depict difference spectra for the enzyme in the presence of oxygen (red) or oxygen and the MbnA substrate peptide (blue).