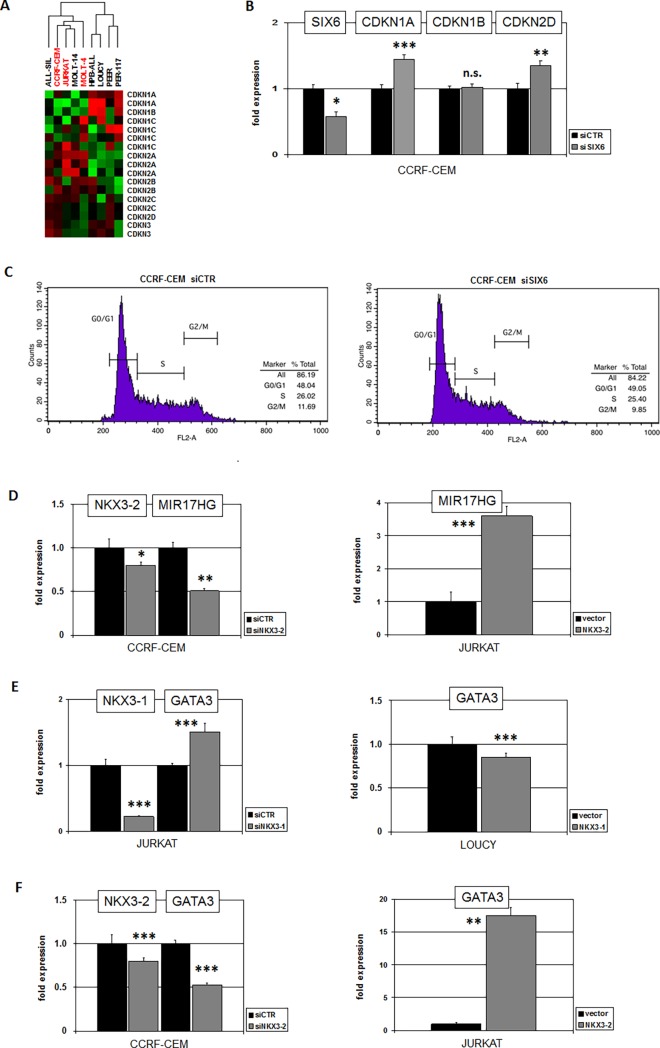
Fig 6. Functional analyses of SIX6 and NKX3-2.
(A) Heatmap showing gene activities in 9 T-ALL cell lines of cyclin-dependent kinase inhibitors. Green represents low and red high expression levels. Cluster analysis showed clustering of SIX6-positive cell lines (red), indicating similarities in expression levels. (B) SiRNA-mediated knockdown of SIX6 in CCRF-CEM resulted in elevated expression of CDKN1A and CDKN2D while CDKN1B remained unchanged. (C) Cell cycle analysis of CCRF-CEM after siRNA-mediated knockdown of SIX6 by flow cytometry indicated no significant differences. (D) SiRNA-mediated knockdown of NKX3-2 in CCRF-CEM resulted in reduced expression (left), and forced expression of NKX3-2 in JURKAT in enhanced expression of MIR17HG (right). These results indicated that NKX3-2 activated MIR17HG expression in T-ALL. (E) SiRNA-mediated knockdown of NKX3-1 in JURKAT resulted in elevated expression (left), and forced expression of NKX3-1 in LOUCY in reduced expression of GATA3 (right). These results indicated that NKX3-1 inhibited GATA3 expression in T-ALL. (F) SiRNA-mediated knockdown of NKX3-2 in CCRF-CEM resulted in reduced expression (left), and forced expression of NKX3-2 in JURKAT in enhanced expression of GATA3 (right). These results indicated that NKX3-2 activated GATA3 expression in T-ALL.