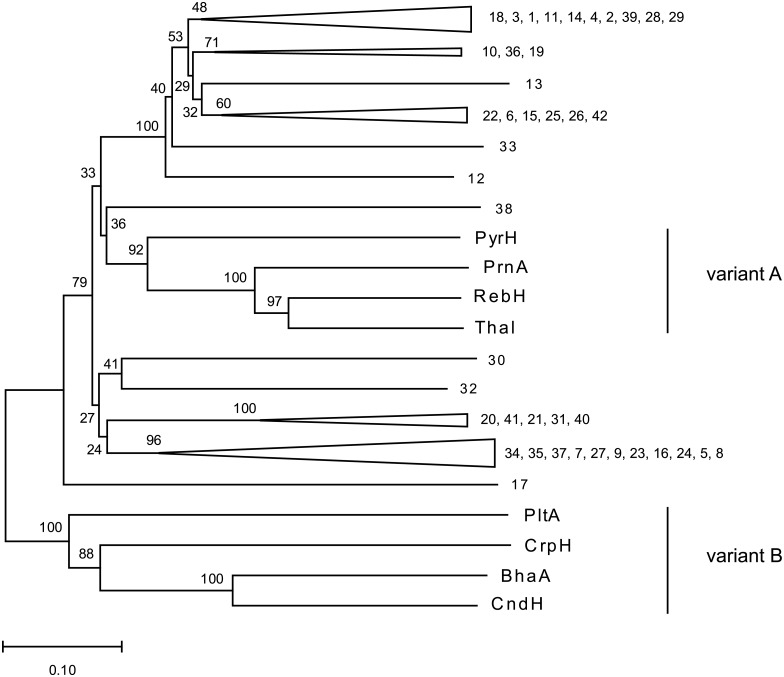
Fig 1. Phylogenetic tree of positive detected FHals from the Botany Bay metagenome in comparison to known FHals on amino acid level.
The tree was constructed using neighbour joining method, bootstrap 1000, with the alignment based on amino acids by MEGA7 [43] and MUSCLE [39]. 1–42: positive FHal hits from the Botany Bay metagenome; PyrH, PrnA, RebH, Thal: Tryptophan halogenases belonging to variant A and accept free substrate; PltA, CrpH, BhaA, CndH: FHals belonging to variant B and require carrier bound substrate. The amino acid sequences are shown in S3 Table.